HAEM5:Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion: Difference between revisions
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) No edit summary |
Bailey.Glen (talk | contribs) No edit summary |
||
| (13 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion}} | {{DISPLAYTITLE:Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion}} | ||
[[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] | ||
{{Under Construction}} | {{Under Construction}} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Acute Myeloid Leukemia (AML) with t(8;21)(q22;q22.1); RUNX1-RUNX1T1]]. | ||
}}</blockquote> | }}</blockquote> | ||
<span style="color:#0070C0">(General Instructions – The focus of these pages is the clinically significant genetic alterations in each disease type. This is based on up-to-date knowledge from multiple resources such as PubMed and the WHO classification books. The CCGA is meant to be a supplemental resource to the WHO classification books; the CCGA captures in a continually updated wiki-stye manner the current genetics/genomics knowledge of each disease, which evolves more rapidly than books can be revised and published. If the same disease is described in multiple WHO classification books, the genetics-related information for that disease will be consolidated into a single main page that has this template (other pages would only contain a link to this main page). Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ <u>HGVS-based nomenclature for variants</u>], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column in a table, click nearby within the table and select the > symbol that appears. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>].)</span> | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
Christine Bryke, MD<br>Beth Israel Deaconess Medical Center, Boston, MA | Christine Bryke, MD<br>Beth Israel Deaconess Medical Center, Boston, MA | ||
==WHO Classification of Disease== | |||
{| class="wikitable" | |||
!Structure | |||
!Disease | |||
|- | |||
|Book | |||
|Haematolymphoid Tumours (5th ed.) | |||
|- | |||
|Category | |||
|Myeloid proliferations and neoplasms | |||
|- | |||
|Family | |||
|Acute myeloid leukaemia | |||
|- | |||
|Type | |||
|Acute myeloid leukaemia with defining genetic abnormalities | |||
|- | |||
|Subtype(s) | |||
|Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion | |||
|} | |||
==Related Terminology== | |||
== | |||
{| class="wikitable" | {| class="wikitable" | ||
| | |+ | ||
| | |Acceptable | ||
|Acute myeloid leukaemia with t(8;21)(q22;q22.1) | |||
|- | |- | ||
| | |Not Recommended | ||
| | |Acute myeloid leukaemia with AML1/ETO | ||
|} | |} | ||
==Gene Rearrangements== | |||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.'')</span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
! | !Driver Gene!!Fusion(s) and Common Partner Genes!!Molecular Pathogenesis!!Typical Chromosomal Alteration(s) | ||
!Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | |||
!Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | |||
!Established Clinical Significance Per Guidelines - Yes or No (Source) | |||
!Clinical Relevance Details/Other Notes | |||
|- | |- | ||
| | |<span class="blue-text">EXAMPLE:</span> ''ABL1''||<span class="blue-text">EXAMPLE:</span> ''BCR::ABL1''||<span class="blue-text">EXAMPLE:</span> The pathogenic derivative is the der(22) resulting in fusion of 5’ BCR and 3’ABL1.||<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2) | ||
|<span class="blue-text">EXAMPLE:</span> Common (CML) | |||
|<span class="blue-text">EXAMPLE:</span> D, P, T | |||
|<span class="blue-text">EXAMPLE:</span> Yes (WHO, NCCN) | |||
|<span class="blue-text">EXAMPLE:</span> | |||
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). BCR::ABL1 is generally favorable in CML (add reference). | |||
|- | |- | ||
| | |<span class="blue-text">EXAMPLE:</span> ''CIC'' | ||
|<span class="blue-text">EXAMPLE:</span> ''CIC::DUX4'' | |||
|<span class="blue-text">EXAMPLE:</span> Typically, the last exon of ''CIC'' is fused to ''DUX4''. The fusion breakpoint in ''CIC'' is usually intra-exonic and removes an inhibitory sequence, upregulating ''PEA3'' genes downstream of ''CIC'' including ''ETV1'', ''ETV4'', and ''ETV5''. | |||
|<span class="blue-text">EXAMPLE:</span> t(4;19)(q25;q13) | |||
|<span class="blue-text">EXAMPLE:</span> Common (CIC-rearranged sarcoma) | |||
|<span class="blue-text">EXAMPLE:</span> D | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> | |||
''DUX4'' has many homologous genes; an alternate translocation in a minority of cases is t(10;19), but this is usually indistinguishable from t(4;19) by short-read sequencing (add references). | |||
|- | |- | ||
| | |<span class="blue-text">EXAMPLE:</span> ''ALK'' | ||
|<span class="blue-text">EXAMPLE:</span> ''ELM4::ALK'' | |||
| | |||
Other fusion partners include ''KIF5B, NPM1, STRN, TFG, TPM3, CLTC, KLC1'' | |||
|<span class="blue-text">EXAMPLE:</span> Fusions result in constitutive activation of the ''ALK'' tyrosine kinase. The most common ''ALK'' fusion is ''EML4::ALK'', with breakpoints in intron 19 of ''ALK''. At the transcript level, a variable (5’) partner gene is fused to 3’ ''ALK'' at exon 20. Rarely, ''ALK'' fusions contain exon 19 due to breakpoints in intron 18. | |||
|<span class="blue-text">EXAMPLE:</span> N/A | |||
|<span class="blue-text">EXAMPLE:</span> Rare (Lung adenocarcinoma) | |||
|<span class="blue-text">EXAMPLE:</span> T | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> | |||
Both balanced and unbalanced forms are observed by FISH (add references). | |||
|- | |- | ||
|<span class="blue-text">EXAMPLE:</span> ''ABL1'' | |||
|<span class="blue-text">EXAMPLE:</span> N/A | |||
|<span class="blue-text">EXAMPLE:</span> Intragenic deletion of exons 2–7 in ''EGFR'' removes the ligand-binding domain, resulting in a constitutively active tyrosine kinase with downstream activation of multiple oncogenic pathways. | |||
|<span class="blue-text">EXAMPLE:</span> N/A | |||
|<span class="blue-text">EXAMPLE:</span> Recurrent (IDH-wildtype Glioblastoma) | |||
|<span class="blue-text">EXAMPLE:</span> D, P, T | |||
| | |||
| | |||
|- | |- | ||
| | | | ||
| | |||
| | | | ||
| | |||
| | |||
| | | | ||
| | |||
| | |||
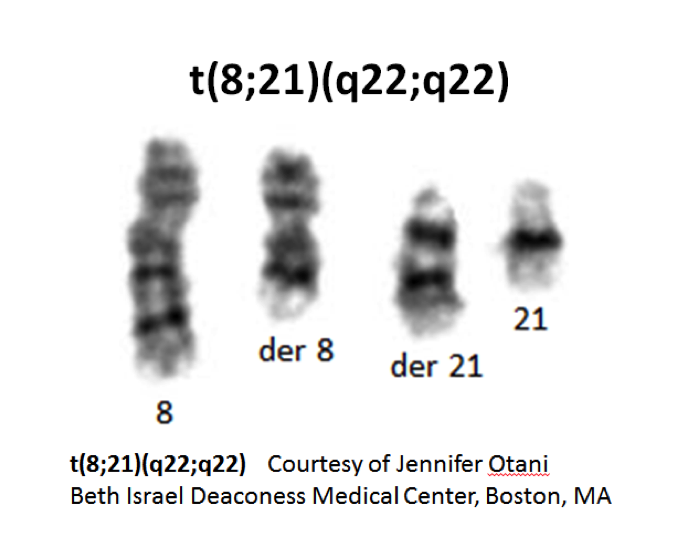
|}[[File:T_8_21_q22_q22.png|frame| Karyogram showing t(8;21)(q22;q22) translocation. Jennifer | |||
Otani, Beth Israel Deaconess Medical Center, Boston, MA|alt=|left]] | |||
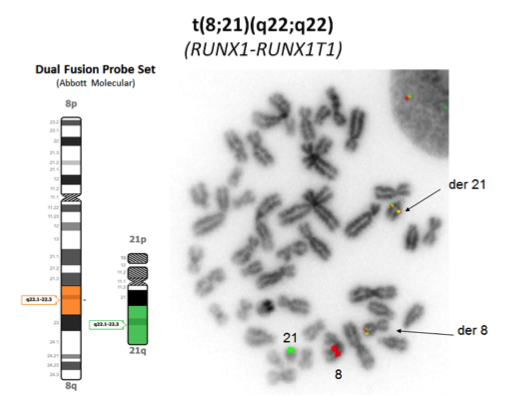
[[File:t(8;21)(q22;q22).png|frame|center| t(8;21)(q22;q22)]] | |||
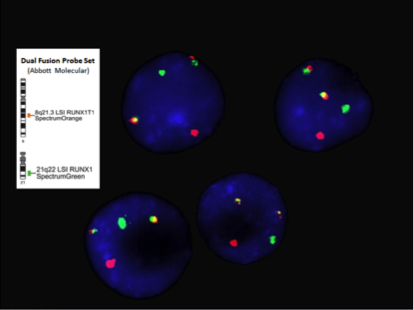
<blockquote class= | [[File:RUNX1_RUNXT1_FISH_Inverted.png|frame|center| FISH RUNX1-RUNX1T1 probe set on metaphase cell with | ||
t(8;21)(q22;q22) translocation. Jennifer Otani, Beth Israel | |||
Deaconess Medical Center, Boston, MA. | |||
]] | |||
<blockquote class="blockedit">{{Box-round|title=v4:Chromosomal Rearrangements (Gene Fusions)|The content below was from the old template. Please incorporate above.}}</blockquote> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
| Line 151: | Line 126: | ||
|} | |} | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).|Please incorporate this section into the relevant tables found in: | ||
* Chromosomal Rearrangements (Gene Fusions) | * Chromosomal Rearrangements (Gene Fusions) | ||
* Individual Region Genomic Gain/Loss/LOH | * Individual Region Genomic Gain/Loss/LOH | ||
* Characteristic Chromosomal Patterns | * Characteristic Chromosomal Patterns | ||
* Gene Mutations (SNV/INDEL)}} | * Gene Mutations (SNV/INDEL)}}</blockquote> | ||
Diagnostic: | Diagnostic: | ||
| Line 176: | Line 154: | ||
*Recently homoharringtonine, aclarubicin and cytarabine as the first course of induction therapy has been found to be highly effective for acute myeloid leukemia with t(8;21)(q22;q22)<ref name=":0" />. | *Recently homoharringtonine, aclarubicin and cytarabine as the first course of induction therapy has been found to be highly effective for acute myeloid leukemia with t(8;21)(q22;q22)<ref name=":0" />. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain/Loss/LOH== | ||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene rearrangements. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Can refer to CGC workgroup tables as linked on the homepage if applicable. Please include references throughout the table. Do not delete the table.'') </span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Chr #!!Gain | !Chr #!!Gain, Loss, Amp, LOH!!Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size]!!Relevant Gene(s) | ||
!Diagnostic | !Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | ||
!Established Clinical Significance Per Guidelines - Yes or No (Source) | |||
! | !Clinical Relevance Details/Other Notes | ||
!Notes | |||
|- | |- | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
7 | 7 | ||
|EXAMPLE Loss | |<span class="blue-text">EXAMPLE:</span> Loss | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
chr7 | chr7 | ||
| | |<span class="blue-text">EXAMPLE:</span> | ||
| | Unknown | ||
|No | |<span class="blue-text">EXAMPLE:</span> D, P | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> No | ||
|<span class="blue-text">EXAMPLE:</span> | |||
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add references). | ||
|- | |- | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
8 | 8 | ||
|EXAMPLE Gain | |<span class="blue-text">EXAMPLE:</span> Gain | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
chr8 | chr8 | ||
| | |<span class="blue-text">EXAMPLE:</span> | ||
| | Unknown | ||
| | |<span class="blue-text">EXAMPLE:</span> D, P | ||
|EXAMPLE | | | ||
|<span class="blue-text">EXAMPLE:</span> | |||
Common recurrent secondary finding for t(8;21) (add | Common recurrent secondary finding for t(8;21) (add references). | ||
|- | |||
|<span class="blue-text">EXAMPLE:</span> | |||
17 | |||
|<span class="blue-text">EXAMPLE:</span> Amp | |||
|<span class="blue-text">EXAMPLE:</span> | |||
17q12; chr17:39,700,064-39,728,658 [hg38; 28.6 kb] | |||
|<span class="blue-text">EXAMPLE:</span> | |||
''ERBB2'' | |||
|<span class="blue-text">EXAMPLE:</span> D, P, T | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> | |||
Amplification of ''ERBB2'' is associated with HER2 overexpression in HER2 positive breast cancer (add references). Add criteria for how amplification is defined. | |||
|- | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Genomic Gain/Loss/LOH|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
9q22 loss | 9q22 loss | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal or Other Global Mutational Patterns== | ||
Put your text here and fill in the table <span style="color:#0070C0">(I''nstructions: Included in this category are alterations such as hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis; microsatellite instability; homologous recombination deficiency; mutational signature pattern; etc. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.'')</span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Chromosomal Pattern | !Chromosomal Pattern | ||
! | !Molecular Pathogenesis | ||
!Prognostic Significance | !Prevalence - | ||
! | Common >20%, Recurrent 5-20% or Rare <5% (Disease) | ||
!Notes | !Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | ||
!Established Clinical Significance Per Guidelines - Yes or No (Source) | |||
!Clinical Relevance Details/Other Notes | |||
|- | |- | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| | |<span class="blue-text">EXAMPLE:</span> See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
|<span class="blue-text">EXAMPLE:</span> Common (Oligodendroglioma) | |||
|<span class="blue-text">EXAMPLE:</span> D, P | |||
| | |||
| | |||
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | |- | ||
|<span class="blue-text">EXAMPLE:</span> | |||
Microsatellite instability - hypermutated | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> Common (Endometrial carcinoma) | |||
|<span class="blue-text">EXAMPLE:</span> P, T | |||
| | |||
| | |||
|- | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Characteristic Chromosomal Aberrations / Patterns|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
Additional aberrations in >70% of cases with t(8;21); common loss of a sex chromosome or deletion 9q | Additional aberrations in >70% of cases with t(8;21); common loss of a sex chromosome or deletion 9q | ||
Trisomy 4 is a rare numerical abnormality detected in 3% of AML patients with t(8;21) (PMIDs: 12682630, 26573082, 27451976). A negative effect on prognosis this group of patients has been attributed to the presence of the mutations in the KIT gene.. Gain of chromosome 4 would likely result in increased dosage of the mutant KIT allele (PMIDs: 16291592, 30899636). | |||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV/INDEL)== | ||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent or common as well either disease defining and/or clinically significant. If a gene has multiple mechanisms depending on the type or site of the alteration, add multiple entries in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Details on clinical significance such as prognosis and other important information such as concomitant and mutually exclusive mutations can be provided in the notes section. Please include references throughout the table. Do not delete the table.'') </span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene | !Gene!!Genetic Alteration!!Tumor Suppressor Gene, Oncogene, Other!!Prevalence - | ||
! | Common >20%, Recurrent 5-20% or Rare <5% (Disease) | ||
!Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | |||
! | !Established Clinical Significance Per Guidelines - Yes or No (Source) | ||
!Notes | !Clinical Relevance Details/Other Notes | ||
|- | |- | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span>''EGFR'' | ||
EXAMPLE: | <br /> | ||
|<span class="blue-text">EXAMPLE:</span> Exon 18-21 activating mutations | |||
|<span class="blue-text">EXAMPLE:</span> Oncogene | |||
|<span class="blue-text">EXAMPLE:</span> Common (lung cancer) | |||
EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> T | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Yes (NCCN) | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Exons 18, 19, and 21 mutations are targetable for therapy. Exon 20 T790M variants cause resistance to first generation TKI therapy and are targetable by second and third generation TKIs (add references). | ||
|- | |||
EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> ''TP53''; Variable LOF mutations | ||
|EXAMPLE: | <br /> | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Variable LOF mutations | ||
|<span class="blue-text">EXAMPLE:</span> Tumor Supressor Gene | |||
|<span class="blue-text">EXAMPLE:</span> Common (breast cancer) | |||
|<span class="blue-text">EXAMPLE:</span> P | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> >90% are somatic; rare germline alterations associated with Li-Fraumeni syndrome (add reference). Denotes a poor prognosis in breast cancer. | |||
|- | |||
|<span class="blue-text">EXAMPLE:</span> ''BRAF''; Activating mutations | |||
|<span class="blue-text">EXAMPLE:</span> Activating mutations | |||
|<span class="blue-text">EXAMPLE:</span> Oncogene | |||
|<span class="blue-text">EXAMPLE:</span> Common (melanoma) | |||
|<span class="blue-text">EXAMPLE:</span> T | |||
| | |||
| | |||
|- | |||
| | |||
| | |||
| | |||
| | |||
| | | | ||
| | | | ||
| | | | ||
|}Note: A more extensive list of mutations can be found in [https://www.cbioportal.org/ <u>cBioportal</u>], [https://cancer.sanger.ac.uk/cosmic <u>COSMIC</u>], and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | |||
|} | |||
Note: A more extensive list of mutations can be found in | |||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Gene Mutations (SNV/INDEL)|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
AML with t(8;21)(q22;q22) has genetic heterogeneity at the molecular level. Several genes have been identified that are frequently mutated in this subset of AML, some of them adversely affecting prognosis. | AML with t(8;21)(q22;q22) has genetic heterogeneity at the molecular level. Several genes have been identified that are frequently mutated in this subset of AML, some of them adversely affecting prognosis. | ||
| Line 324: | Line 351: | ||
|} | |} | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Epigenomic Alterations== | ==Epigenomic Alterations== | ||
| Line 331: | Line 361: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: | |||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Please include references throughout the table. Do not delete the table.)''</span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
|EXAMPLE: BRAF and MAP2K1; Activating mutations | |<span class="blue-text">EXAMPLE:</span> ''BRAF'' and ''MAP2K1''; Activating mutations | ||
|EXAMPLE: MAPK signaling | |<span class="blue-text">EXAMPLE:</span> MAPK signaling | ||
|EXAMPLE: Increased cell growth and proliferation | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation | ||
|- | |- | ||
|EXAMPLE: CDKN2A; Inactivating mutations | |<span class="blue-text">EXAMPLE:</span> ''CDKN2A''; Inactivating mutations | ||
|EXAMPLE: Cell cycle regulation | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation | ||
|EXAMPLE: Unregulated cell division | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division | ||
|- | |- | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> ''KMT2C'' and ''ARID1A''; Inactivating mutations | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program | ||
|- | |||
| | |||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Genes and Main Pathways Involved|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
Genes: | Genes: | ||
| Line 372: | Line 407: | ||
*The normal transcriptional activity of the RUNX1 protein in the core binding factor is affected in a negative manner by the chimeric protein encoded by the RUNX1-RUNX1T1 fusion gene. It has been proposed that the RUNX1-RUNX1T1 protein recruits histone deacetylases and DNA methyl transferase 1 to core binding factor target genes<ref>{{Cite journal|last=Marcucci|first=Guido|date=2006|title=Core binding factor acute myeloid leukemia|url=https://www.ncbi.nlm.nih.gov/pubmed/16728942|journal=Clinical Advances in Hematology & Oncology: H&O|volume=4|issue=5|pages=339–341|issn=1543-0790|pmid=16728942}}</ref>. This leads to increased chromatin deacetylation and promotor hypermethylation, resulting in gene transcriptional repression and disruption of normal cellular pathways of hematopoiesis. | *The normal transcriptional activity of the RUNX1 protein in the core binding factor is affected in a negative manner by the chimeric protein encoded by the RUNX1-RUNX1T1 fusion gene. It has been proposed that the RUNX1-RUNX1T1 protein recruits histone deacetylases and DNA methyl transferase 1 to core binding factor target genes<ref>{{Cite journal|last=Marcucci|first=Guido|date=2006|title=Core binding factor acute myeloid leukemia|url=https://www.ncbi.nlm.nih.gov/pubmed/16728942|journal=Clinical Advances in Hematology & Oncology: H&O|volume=4|issue=5|pages=339–341|issn=1543-0790|pmid=16728942}}</ref>. This leads to increased chromatin deacetylation and promotor hypermethylation, resulting in gene transcriptional repression and disruption of normal cellular pathways of hematopoiesis. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Genetic Diagnostic Testing Methods== | ==Genetic Diagnostic Testing Methods== | ||
| Line 389: | Line 427: | ||
==Additional Information== | ==Additional Information== | ||
This disease is <u>defined/characterized</u> as detailed below: | |||
Acute myeloid leukemia (AML) with t(8;21)(q22;q22) resulting in RUNX1-RUNX1T1 gene fusion defines a subtype of AML with neutrophil lineage maturation. In the 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia, it is in the group of AML with recurrent genetic abnormalities<ref>{{Cite journal|last=Arber|first=Daniel A.|last2=Orazi|first2=Attilio|last3=Hasserjian|first3=Robert|last4=Thiele|first4=Jürgen|last5=Borowitz|first5=Michael J.|last6=Le Beau|first6=Michelle M.|last7=Bloomfield|first7=Clara D.|last8=Cazzola|first8=Mario|last9=Vardiman|first9=James W.|date=2016|title=The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia|url=https://www.ncbi.nlm.nih.gov/pubmed/27069254|journal=Blood|volume=127|issue=20|pages=2391–2405|doi=10.1182/blood-2016-03-643544|issn=1528-0020|pmid=27069254}}</ref>. In the older French-American-British (FAB) classification system, t(8;21)(q22;q22) positive AML belonged to the acute myeloblastic leukemia with maturation M2 category and accounted for 10% of cases. t(8;21)(q22;q22) can occasionally be seen in therapy-related AML. In those instances the leukemia is not classified in the group of AML with recurrent genetic abnormalities due to clinical and prognostic differences. AML with t(8;21)(q22;q22) is a core binding leukemia (see Genes and Main Pathways Involved section below)<ref>{{Cite journal|last=Sangle|first=Nikhil A.|last2=Perkins|first2=Sherrie L.|date=2011|title=Core-binding factor acute myeloid leukemia|url=https://www.ncbi.nlm.nih.gov/pubmed/22032582|journal=Archives of Pathology & Laboratory Medicine|volume=135|issue=11|pages=1504–1509|doi=10.5858/arpa.2010-0482-RS|issn=1543-2165|pmid=22032582}}</ref>. | |||
*t(8;21)(q22;q22) is a translocation involving the long arms of chromosomes 8 and 21 that defines a distinctive cytogenetic subtype of [[AML|acute myeloid leukemia]]. | |||
*Complex translocations involving chromosomes 8, 21 and a third chromosome occur; the derivative chromosome 8 in the t(8;21)(q22;q22) and the variant t(8;21;var) carries the RUNX1-RUNX1T1 gene rearrangement. | |||
*Cases with cryptic RUNX1-RUNX1T1 gene rearrangement have been reported. | |||
*Note: The RUNX1T1 gene is currently located at 8q21.3. This gene was previously located at 8q22, which explains the discrepancy in the breakpoint given in the name for this WHO-defined disease entity. | |||
The <u>epidemiology/prevalence</u> of this disease is detailed below: | |||
Approximately 1-5% of AML cases have the t(8;21)(q22;q22.1) | |||
*Occurs mainly in younger patients | |||
*Mean age is 30 years | |||
*Most common type of AML in children | |||
The <u>clinical features</u> of this disease are detailed below: | |||
*May present as a myeloid sarcoma and, if so, the bone marrow aspirate blast count may be misleadingly low | |||
*Concurrent systemic mastocytosis has been reported | |||
The <u>sites of involvement</u> of this disease are detailed below: | |||
*Bone marrow | |||
The <u>morphologic features</u> of this disease are detailed below: | |||
*Large blasts with abundant basophilic cytoplasm often with azurophilic granules and perinuclear clearing | |||
*Long sharp Auer rods are frequently observed. | |||
*Promyelocytes, myelocytes and metamyelocytes may have abnormal nuclear segmentation and cytoplasmic staining. | |||
*Normal eosinophil precursors are frequently increased and do not show the nuclear and cytoplasmic abnormalities seen in AML with inv(16)(p13.1q22) or t(16;16)(p13.1;q22). | |||
*Cases with t(8;21)(q22;q22.1) and less than 20% blast percentage in the marrow should be classified as AML rather than myelodysplastic syndrome according to the WHO 2017<ref>Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France.</ref>. | |||
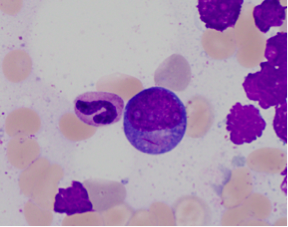
[[File:AML_RUNX1_RUNXT1_Path.png|center|frame|Bone marrow aspirate with t(8;21)(q22;q22) | |||
blast with Auer rod (100X). Robert Willim, MD, | |||
Beth Israel Deaconess Medical Center, Boston, MA]]The <u>immunophenotype</u> of this disease is detailed below: | |||
*Blasts express CD34, HLA-DR, MPO, CD13, CD33 (weak), CD15 and/or CD65, co-expression of CD34. Lymphoid markers: CD19, PAX5, sometimes cytoplasmic CD79a | |||
*Occasional weak TdT positivity and/or CD56 expression may have adverse prognosis | |||
==Links== | ==Links== | ||
| Line 406: | Line 484: | ||
==References== | ==References== | ||
(use the "Cite" icon at the top of the page) <span style="color:#0070C0">(''Instructions: Add each reference into the text above by clicking | (use the "Cite" icon at the top of the page) <span style="color:#0070C0">(''Instructions: Add each reference into the text above by clicking where you want to insert the reference, selecting the “Cite” icon at the top of the wiki page, and using the “Automatic” tab option to search by PMID to select the reference to insert. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference. To insert the same reference again later in the page, select the “Cite” icon and “Re-use” to find the reference; DO NOT insert the same reference twice using the “Automatic” tab as it will be treated as two separate references. The reference list in this section will be automatically generated and sorted''</span><span style="color:#0070C0">''.''</span><span style="color:#0070C0">)</span> <references /> | ||
<br /> | |||
==Notes== | ==Notes== | ||
| Line 416: | Line 494: | ||
<nowiki>*</nowiki>''Citation of this Page'': “Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_RUNX1::RUNX1T1_fusion</nowiki>. | <nowiki>*</nowiki>''Citation of this Page'': “Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_RUNX1::RUNX1T1_fusion</nowiki>. | ||
[[Category:HAEM5]][[Category:DISEASE]][[Category:Diseases A]] | [[Category:HAEM5]] | ||
[[Category:DISEASE]] | |||
[[Category:Diseases A]] | |||
Latest revision as of 12:08, 3 July 2025
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Acute Myeloid Leukemia (AML) with t(8;21)(q22;q22.1); RUNX1-RUNX1T1.
(General Instructions – The focus of these pages is the clinically significant genetic alterations in each disease type. This is based on up-to-date knowledge from multiple resources such as PubMed and the WHO classification books. The CCGA is meant to be a supplemental resource to the WHO classification books; the CCGA captures in a continually updated wiki-stye manner the current genetics/genomics knowledge of each disease, which evolves more rapidly than books can be revised and published. If the same disease is described in multiple WHO classification books, the genetics-related information for that disease will be consolidated into a single main page that has this template (other pages would only contain a link to this main page). Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column in a table, click nearby within the table and select the > symbol that appears. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support.)
Primary Author(s)*
Christine Bryke, MD
Beth Israel Deaconess Medical Center, Boston, MA
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Acute myeloid leukaemia |
| Type | Acute myeloid leukaemia with defining genetic abnormalities |
| Subtype(s) | Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion |
Related Terminology
| Acceptable | Acute myeloid leukaemia with t(8;21)(q22;q22.1) |
| Not Recommended | Acute myeloid leukaemia with AML1/ETO |
Gene Rearrangements
Put your text here and fill in the table (Instructions: Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Driver Gene | Fusion(s) and Common Partner Genes | Molecular Pathogenesis | Typical Chromosomal Alteration(s) | Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: ABL1 | EXAMPLE: BCR::ABL1 | EXAMPLE: The pathogenic derivative is the der(22) resulting in fusion of 5’ BCR and 3’ABL1. | EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: Common (CML) | EXAMPLE: D, P, T | EXAMPLE: Yes (WHO, NCCN) | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). BCR::ABL1 is generally favorable in CML (add reference). |
| EXAMPLE: CIC | EXAMPLE: CIC::DUX4 | EXAMPLE: Typically, the last exon of CIC is fused to DUX4. The fusion breakpoint in CIC is usually intra-exonic and removes an inhibitory sequence, upregulating PEA3 genes downstream of CIC including ETV1, ETV4, and ETV5. | EXAMPLE: t(4;19)(q25;q13) | EXAMPLE: Common (CIC-rearranged sarcoma) | EXAMPLE: D | EXAMPLE:
DUX4 has many homologous genes; an alternate translocation in a minority of cases is t(10;19), but this is usually indistinguishable from t(4;19) by short-read sequencing (add references). | |
| EXAMPLE: ALK | EXAMPLE: ELM4::ALK
|
EXAMPLE: Fusions result in constitutive activation of the ALK tyrosine kinase. The most common ALK fusion is EML4::ALK, with breakpoints in intron 19 of ALK. At the transcript level, a variable (5’) partner gene is fused to 3’ ALK at exon 20. Rarely, ALK fusions contain exon 19 due to breakpoints in intron 18. | EXAMPLE: N/A | EXAMPLE: Rare (Lung adenocarcinoma) | EXAMPLE: T | EXAMPLE:
Both balanced and unbalanced forms are observed by FISH (add references). | |
| EXAMPLE: ABL1 | EXAMPLE: N/A | EXAMPLE: Intragenic deletion of exons 2–7 in EGFR removes the ligand-binding domain, resulting in a constitutively active tyrosine kinase with downstream activation of multiple oncogenic pathways. | EXAMPLE: N/A | EXAMPLE: Recurrent (IDH-wildtype Glioblastoma) | EXAMPLE: D, P, T | ||

editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence |
|---|---|---|---|
| t(8;21)(q22;q22.1) | 5' RUNX1 / 3'RUNX1T1 | der(8) | 1- 5% of AML |
End of V4 Section
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
Diagnostic:
- The presence of t(8;21)(q22;q22) is diagnostic of a cytogenetic subtype of acute myeloid leukemia with maturation in the neutrophilic lineage.
- When t(8;21)(q22;q22) is present the diagnosis of acute myeloid leukemia is made regardless of a blast count less than 20%.
Prognostic:
- t(8;21)(q22;q22) positive AML usually has a good response to chemotherapy and a high complete remission rate with long-term disease-free survival when treated with high dose cytarabine in the consolidation phase. Factors that appear to adversely affected prognosis include CD56 expression and KIT gene mutations. The prognostic significance of other accompanying gene mutations is under investigation.
Therapeutic target:
- t(8;21)(q22;q22) positive AML currently has no molecular target for drug therapy.
- Standard induction 7 + 3 chemotherapy regimen of cytarabine for 7 days plus an anthracycline or anthracenedione for 3 days. In first complete remission patients younger than 60 years without a KIT mutation are treated with high dose cytarabine consolidation therapy. If a KIT mutation is present, bone marrow transplant is performed in first complete remission[1].
- Recently homoharringtonine, aclarubicin and cytarabine as the first course of induction therapy has been found to be highly effective for acute myeloid leukemia with t(8;21)(q22;q22)[2].
End of V4 Section
Individual Region Genomic Gain/Loss/LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene rearrangements. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Can refer to CGC workgroup tables as linked on the homepage if applicable. Please include references throughout the table. Do not delete the table.)
| Chr # | Gain, Loss, Amp, LOH | Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size] | Relevant Gene(s) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7 |
EXAMPLE:
Unknown |
EXAMPLE: D, P | EXAMPLE: No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add references). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8 |
EXAMPLE:
Unknown |
EXAMPLE: D, P | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add references). | |
| EXAMPLE:
17 |
EXAMPLE: Amp | EXAMPLE:
17q12; chr17:39,700,064-39,728,658 [hg38; 28.6 kb] |
EXAMPLE:
ERBB2 |
EXAMPLE: D, P, T | EXAMPLE:
Amplification of ERBB2 is associated with HER2 overexpression in HER2 positive breast cancer (add references). Add criteria for how amplification is defined. | |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.
9q22 loss
End of V4 Section
Characteristic Chromosomal or Other Global Mutational Patterns
Put your text here and fill in the table (Instructions: Included in this category are alterations such as hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis; microsatellite instability; homologous recombination deficiency; mutational signature pattern; etc. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Chromosomal Pattern | Molecular Pathogenesis | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
EXAMPLE: See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | EXAMPLE: Common (Oligodendroglioma) | EXAMPLE: D, P | ||
| EXAMPLE:
Microsatellite instability - hypermutated |
EXAMPLE: Common (Endometrial carcinoma) | EXAMPLE: P, T | |||
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.
Additional aberrations in >70% of cases with t(8;21); common loss of a sex chromosome or deletion 9q
Trisomy 4 is a rare numerical abnormality detected in 3% of AML patients with t(8;21) (PMIDs: 12682630, 26573082, 27451976). A negative effect on prognosis this group of patients has been attributed to the presence of the mutations in the KIT gene.. Gain of chromosome 4 would likely result in increased dosage of the mutant KIT allele (PMIDs: 16291592, 30899636).
End of V4 Section
Gene Mutations (SNV/INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent or common as well either disease defining and/or clinically significant. If a gene has multiple mechanisms depending on the type or site of the alteration, add multiple entries in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Details on clinical significance such as prognosis and other important information such as concomitant and mutually exclusive mutations can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Gene | Genetic Alteration | Tumor Suppressor Gene, Oncogene, Other | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
| EXAMPLE:EGFR
|
EXAMPLE: Exon 18-21 activating mutations | EXAMPLE: Oncogene | EXAMPLE: Common (lung cancer) | EXAMPLE: T | EXAMPLE: Yes (NCCN) | EXAMPLE: Exons 18, 19, and 21 mutations are targetable for therapy. Exon 20 T790M variants cause resistance to first generation TKI therapy and are targetable by second and third generation TKIs (add references). |
| EXAMPLE: TP53; Variable LOF mutations
|
EXAMPLE: Variable LOF mutations | EXAMPLE: Tumor Supressor Gene | EXAMPLE: Common (breast cancer) | EXAMPLE: P | EXAMPLE: >90% are somatic; rare germline alterations associated with Li-Fraumeni syndrome (add reference). Denotes a poor prognosis in breast cancer. | |
| EXAMPLE: BRAF; Activating mutations | EXAMPLE: Activating mutations | EXAMPLE: Oncogene | EXAMPLE: Common (melanoma) | EXAMPLE: T | ||
Note: A more extensive list of mutations can be found in cBioportal, COSMIC, and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.
AML with t(8;21)(q22;q22) has genetic heterogeneity at the molecular level. Several genes have been identified that are frequently mutated in this subset of AML, some of them adversely affecting prognosis.
- KIT mutations occur in 20-25% of cases and are associated with a high risk of relapse in AML with t(8;21)(q22;q22)[3]. Patients with KIT mutations have a significantly shorter overall survival and disease free survival periods than those without KIT mutations[4].
- In a study of 139 t(8;21)(q22;q22) AML patients were investigated for possible mutations in KIT and 10 other genes (ASXL1, FLT3, NPM1, MLL, IDH1, IDH2, KRAS, NRAS, CBL, and JAK2)[2]. 49.6% had one mutation in addition to the t(8;21)(q22;q22). These were most often KIT, NRAS and ASXL1. 16.5% of patients had ≥2 mutations. Infrequent accompanying mutated genes found in only 2.9-5% of cases include in decreasing frequency: FLT3-ITD, FLT3-TKD, CBL, and KRAS, IDH2 and JAK2. These mutations did not adversely affect prognosis. NPM1 mutations are thought to be mutually exclusive of RUNX1-RUNX1T1.
- ASXL2 mutations have been identified in greater than 20% of patients with t(8;21), but not in patients with inv(16)/t(16;16) or RUNX1-mutated AML[5]. ASXL2 mutations are similarly frequent in adults and children with t(8;21) and are mutually exclusive with ASXL1 mutations. Although overall survival is similar between ASXL1 and ASXL2 mutant t(8;21) AML patients and their wild-type counterparts, patients with ASXL1 or ASXL2 mutations had a cumulative incidence of relapse that is significantly higher than with their ASXL1 or ASXL2 wild-type counterparts[5].
- Recurring ZBTB7A mutations have been identified in 23% (13/56) of AML t(8;21) patients, including missense and truncating mutations resulting in alteration or loss of the C-terminal zinc-finger domain of ZBTB7A[6]. The transcription factor ZBTB7A is important for hematopoietic lineage fate decisions and for regulation of glycolysis. On a functional level, it has been shown that ZBTB7A mutations disrupt the transcriptional repressor potential and the anti-proliferative effect of ZBTB7A. The specific association of ZBTB7A mutations with t(8;21) rearranged AML points towards leukemogenic cooperativity between mutant ZBTB7A and the RUNX1/RUNX1T1 fusion[6].
- WT1 mutations have been found to be relatively common (13.8%) in core binding factor leukemia[7]. Low WT1 transcript levels at diagnosis predicted poor outcomes of acute myeloid leukemia patients with t(8;21) who received chemotherapy or allogeneic hematopoietic stem cell transplantation[8].
| Gene | Mutation | Oncogene/Tumor Suppressor/Other | Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) | Prevalence (COSMIC/TCGA/Other) |
|---|---|---|---|---|
| KIT | 20-30% |
Other Mutations
| Type | Gene/Region/Other |
|---|---|
| Concomitant Mutations | NRAS and ASXL1 |
| Secondary Mutations | |
| Mutually Exclusive | ASXL1 and ASXL2 |
End of V4 Section
Epigenomic Alterations
None
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Please include references throughout the table. Do not delete the table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.
Genes:
- RUNX1T1 (also known as ETO) on 8q22 is transcribed from telomere to centromere; 3 proline rich domains; 2 zinc finger, and a PEST region in the C-terminus; tissue restricted expression; nuclear localization; putative transcription factor.
- RUNX1 (also known as AML1 and CBFA) on 21q22; contains a runt domain and a transactivation domain in the C-terminus; forms heterodimers; widely expressed; nuclear localization; transcription factor (activator) for various hematopoietic-specific genes.
DNA:
- t(8;21)(q22;q22) results in a 5’RUNX1-3’RUNX1T1 hybrid gene involving fusion of the very 5’ end of the RUNX1T1 gene to RUNX1 in an ~25 kb breakpoint region between exons 5 and 6.
Protein:
- A fusion protein with the N-terminus runt domain (responsible for DNA binding and protein-protein interaction) of RUNX1 fused to the 577 C-terminal residues of RUNX1T1. The reciprocal product is not detected. The fusion protein retains the ability to recognize the RUNX1 consensus DNA binding site (it is a negative dominant competitor with the normal RUNX1) and to dimerize with the CBFβ subunit in the heterodimeric core binding factor. The core binding factor is a transcription factor that plays an essential role in regulation of normal hematopoiesis. It is composed of a DNA-binding CBFα (RUNX1) chain and a non-DNA-binding CBFβ chain. It is likely oncogenic due to altered transcriptional regulation of normal RUNX1 target genes. Animal studies suggest that the fusion proteins alone are not able to induce leukemia and that additional genetic alterations are required for leukemogenic transformation[9][10].
Key cellular pathways:
- The normal transcriptional activity of the RUNX1 protein in the core binding factor is affected in a negative manner by the chimeric protein encoded by the RUNX1-RUNX1T1 fusion gene. It has been proposed that the RUNX1-RUNX1T1 protein recruits histone deacetylases and DNA methyl transferase 1 to core binding factor target genes[11]. This leads to increased chromatin deacetylation and promotor hypermethylation, resulting in gene transcriptional repression and disruption of normal cellular pathways of hematopoiesis.
End of V4 Section
Genetic Diagnostic Testing Methods
- Chromosome analysis, FISH, RT-PCR
- FISH or RT-PCR is needed for cytogenetically cryptic cases with typical blast morphology and immunophenotype
- RT-PCR is employed for minimal residual disease detection
Familial Forms
None
Additional Information
This disease is defined/characterized as detailed below:
Acute myeloid leukemia (AML) with t(8;21)(q22;q22) resulting in RUNX1-RUNX1T1 gene fusion defines a subtype of AML with neutrophil lineage maturation. In the 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia, it is in the group of AML with recurrent genetic abnormalities[12]. In the older French-American-British (FAB) classification system, t(8;21)(q22;q22) positive AML belonged to the acute myeloblastic leukemia with maturation M2 category and accounted for 10% of cases. t(8;21)(q22;q22) can occasionally be seen in therapy-related AML. In those instances the leukemia is not classified in the group of AML with recurrent genetic abnormalities due to clinical and prognostic differences. AML with t(8;21)(q22;q22) is a core binding leukemia (see Genes and Main Pathways Involved section below)[13].
- t(8;21)(q22;q22) is a translocation involving the long arms of chromosomes 8 and 21 that defines a distinctive cytogenetic subtype of acute myeloid leukemia.
- Complex translocations involving chromosomes 8, 21 and a third chromosome occur; the derivative chromosome 8 in the t(8;21)(q22;q22) and the variant t(8;21;var) carries the RUNX1-RUNX1T1 gene rearrangement.
- Cases with cryptic RUNX1-RUNX1T1 gene rearrangement have been reported.
- Note: The RUNX1T1 gene is currently located at 8q21.3. This gene was previously located at 8q22, which explains the discrepancy in the breakpoint given in the name for this WHO-defined disease entity.
The epidemiology/prevalence of this disease is detailed below:
Approximately 1-5% of AML cases have the t(8;21)(q22;q22.1)
- Occurs mainly in younger patients
- Mean age is 30 years
- Most common type of AML in children
The clinical features of this disease are detailed below:
- May present as a myeloid sarcoma and, if so, the bone marrow aspirate blast count may be misleadingly low
- Concurrent systemic mastocytosis has been reported
The sites of involvement of this disease are detailed below:
- Bone marrow
The morphologic features of this disease are detailed below:
- Large blasts with abundant basophilic cytoplasm often with azurophilic granules and perinuclear clearing
- Long sharp Auer rods are frequently observed.
- Promyelocytes, myelocytes and metamyelocytes may have abnormal nuclear segmentation and cytoplasmic staining.
- Normal eosinophil precursors are frequently increased and do not show the nuclear and cytoplasmic abnormalities seen in AML with inv(16)(p13.1q22) or t(16;16)(p13.1;q22).
- Cases with t(8;21)(q22;q22.1) and less than 20% blast percentage in the marrow should be classified as AML rather than myelodysplastic syndrome according to the WHO 2017[14].
The immunophenotype of this disease is detailed below:
- Blasts express CD34, HLA-DR, MPO, CD13, CD33 (weak), CD15 and/or CD65, co-expression of CD34. Lymphoid markers: CD19, PAX5, sometimes cytoplasmic CD79a
- Occasional weak TdT positivity and/or CD56 expression may have adverse prognosis
Links
KIT mutations in AML COSMIC database
www.uptodate.com cytogenetics in AML
www.genenames.org RUNX1
www.genenames.org RUNX1T1
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking where you want to insert the reference, selecting the “Cite” icon at the top of the wiki page, and using the “Automatic” tab option to search by PMID to select the reference to insert. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference. To insert the same reference again later in the page, select the “Cite” icon and “Re-use” to find the reference; DO NOT insert the same reference twice using the “Automatic” tab as it will be treated as two separate references. The reference list in this section will be automatically generated and sorted.)
- ↑ Tallman, Martin S.; et al. (2019-06-01). "Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology". Journal of the National Comprehensive Cancer Network: JNCCN. 17 (6): 721–749. doi:10.6004/jnccn.2019.0028. ISSN 1540-1413. PMID 31200351.
- ↑ 2.0 2.1 Krauth, M.-T.; et al. (2014). "High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome". Leukemia. 28 (7): 1449–1458. doi:10.1038/leu.2014.4. ISSN 1476-5551. PMID 24402164.
- ↑ Paschka, Peter; et al. (2006). "Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study". Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 24 (24): 3904–3911. doi:10.1200/JCO.2006.06.9500. ISSN 1527-7755. PMID 16921041.
- ↑ Kim, Hee-Jin; et al. (2013). "KIT D816 mutation associates with adverse outcomes in core binding factor acute myeloid leukemia, especially in the subgroup with RUNX1/RUNX1T1 rearrangement". Annals of Hematology. 92 (2): 163–171. doi:10.1007/s00277-012-1580-5. ISSN 1432-0584. PMID 23053179.
- ↑ 5.0 5.1 Micol, Jean-Baptiste; et al. (2014). "Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations". Blood. 124 (9): 1445–1449. doi:10.1182/blood-2014-04-571018. ISSN 1528-0020. PMC 4148766. PMID 24973361.
- ↑ 6.0 6.1 Hartmann, Luise; et al. (2016). "ZBTB7A mutations in acute myeloid leukaemia with t(8;21) translocation". Nature Communications. 7: 11733. doi:10.1038/ncomms11733. ISSN 2041-1723. PMC 4895769. PMID 27252013.
- ↑ Park, Sang Hyuk; et al. (2015). "Incidences and Prognostic Impact of c-KIT, WT1, CEBPA, and CBL Mutations, and Mutations Associated With Epigenetic Modification in Core Binding Factor Acute Myeloid Leukemia: A Multicenter Study in a Korean Population". Annals of Laboratory Medicine. 35 (3): 288–297. doi:10.3343/alm.2015.35.3.288. ISSN 2234-3814. PMC 4390696. PMID 25932436.
- ↑ Qin, Ya-Zhen; et al. (2016). "Low WT1 transcript levels at diagnosis predicted poor outcomes of acute myeloid leukemia patients with t(8;21) who received chemotherapy or allogeneic hematopoietic stem cell transplantation". Chinese Journal of Cancer. 35: 46. doi:10.1186/s40880-016-0110-6. ISSN 1944-446X. PMC 4873994. PMID 27197573.
- ↑ Speck, Nancy A.; et al. (2002). "Core-binding factors in haematopoiesis and leukaemia". Nature Reviews. Cancer. 2 (7): 502–513. doi:10.1038/nrc840. ISSN 1474-175X. PMID 12094236.
- ↑ Downing, James R. (2003). "The core-binding factor leukemias: lessons learned from murine models". Current Opinion in Genetics & Development. 13 (1): 48–54. doi:10.1016/s0959-437x(02)00018-7. ISSN 0959-437X. PMID 12573435.
- ↑ Marcucci, Guido (2006). "Core binding factor acute myeloid leukemia". Clinical Advances in Hematology & Oncology: H&O. 4 (5): 339–341. ISSN 1543-0790. PMID 16728942.
- ↑ Arber, Daniel A.; et al. (2016). "The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia". Blood. 127 (20): 2391–2405. doi:10.1182/blood-2016-03-643544. ISSN 1528-0020. PMID 27069254.
- ↑ Sangle, Nikhil A.; et al. (2011). "Core-binding factor acute myeloid leukemia". Archives of Pathology & Laboratory Medicine. 135 (11): 1504–1509. doi:10.5858/arpa.2010-0482-RS. ISSN 1543-2165. PMID 22032582.
- ↑ Arber DA, et al., (2017). Acute myeloid leukaemia with recurrent genetic abnormalities, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, and Siebert R, Editors. IARC Press: Lyon, France.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.
Edited by: Fabiola Quintero-Rivera 7/21/2018
*Citation of this Page: “Acute myeloid leukaemia with RUNX1::RUNX1T1 fusion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 07/3/2025, https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_RUNX1::RUNX1T1_fusion.