HAEM5:Myelodysplastic neoplasm with low blasts and 5q deletion: Difference between revisions
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) No edit summary |
Bailey.Glen (talk | contribs) No edit summary |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:Myelodysplastic neoplasm with low blasts and 5q deletion}} | {{DISPLAYTITLE:Myelodysplastic neoplasm with low blasts and 5q deletion}} | ||
[[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (WHO Classification, 5th ed.)]] | ||
{{Under Construction}} | {{Under Construction}} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=Content Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition Classification|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Myelodysplastic Syndrome (MDS) with Isolated del(5q)]]. | ||
}}</blockquote> | }}</blockquote> | ||
<span style="color:#0070C0">(General Instructions – The focus of these pages is the clinically significant genetic alterations in each disease type. This is based on up-to-date knowledge from multiple resources such as PubMed and the WHO classification books. The CCGA is meant to be a supplemental resource to the WHO classification books; the CCGA captures in a continually updated wiki-stye manner the current genetics/genomics knowledge of each disease, which evolves more rapidly than books can be revised and published. If the same disease is described in multiple WHO classification books, the genetics-related information for that disease will be consolidated into a single main page that has this template (other pages would only contain a link to this main page). Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ <u>HGVS-based nomenclature for variants</u>], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column in a table, click nearby within the table and select the > symbol that appears. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>].)</span> | |||
==Primary Author(s)*== | ==Primary Author(s)*== | ||
Xiaolin Hu, Ph.D; Teresa Smolarek, Ph.D, FACMG | Xiaolin Hu, Ph.D; Teresa Smolarek, Ph.D, FACMG | ||
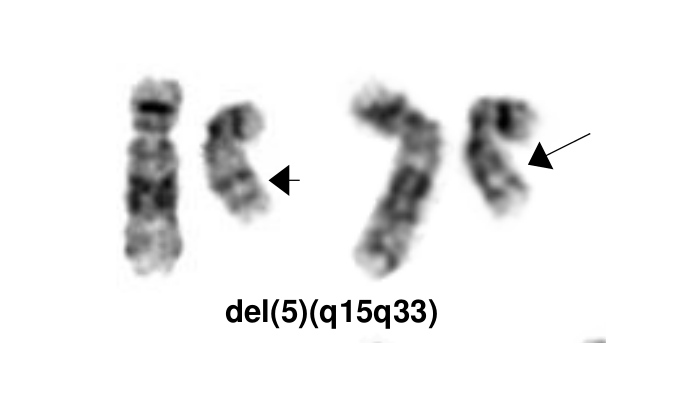
[[File:del(5)(q15q33).png|del(5)(q15q33)|322x322px]] | [[File:del(5)(q15q33).png|del(5)(q15q33)|322x322px]] | ||
== | ==WHO Classification of Disease== | ||
{| class="wikitable" | |||
!Structure | |||
!Disease | |||
|- | |||
|Book | |||
|Haematolymphoid Tumours (5th ed.) | |||
|- | |||
|Category | |||
|Myeloid proliferations and neoplasms | |||
|- | |||
|Family | |||
|Myelodysplastic neoplasms | |||
Myelodysplastic | |- | ||
|Type | |||
|Myelodysplastic neoplasms, with defining genetic abnormalities | |||
|- | |||
|Subtype(s) | |||
|Myelodysplastic neoplasm with low blasts and 5q deletion | |||
|} | |||
== | ==Related Terminology== | ||
{| class="wikitable" | {| class="wikitable" | ||
| | |+ | ||
| | |Acceptable | ||
|Myelodysplastic syndrome with del(5q) | |||
|- | |- | ||
| | |Not Recommended | ||
| | |5q− syndrome | ||
|} | |} | ||
==Gene Rearrangements== | |||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.'')</span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
! | !Driver Gene!!Fusion(s) and Common Partner Genes!!Molecular Pathogenesis!!Typical Chromosomal Alteration(s) | ||
!Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | |||
!Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | |||
!Established Clinical Significance Per Guidelines - Yes or No (Source) | |||
!Clinical Relevance Details/Other Notes | |||
|- | |- | ||
| | |<span class="blue-text">EXAMPLE:</span> ''ABL1''||<span class="blue-text">EXAMPLE:</span> ''BCR::ABL1''||<span class="blue-text">EXAMPLE:</span> The pathogenic derivative is the der(22) resulting in fusion of 5’ BCR and 3’ABL1.||<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2) | ||
|<span class="blue-text">EXAMPLE:</span> Common (CML) | |||
|<span class="blue-text">EXAMPLE:</span> D, P, T | |||
|<span class="blue-text">EXAMPLE:</span> Yes (WHO, NCCN) | |||
|<span class="blue-text">EXAMPLE:</span> | |||
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). BCR::ABL1 is generally favorable in CML (add reference). | |||
|- | |- | ||
| | |<span class="blue-text">EXAMPLE:</span> ''CIC'' | ||
|- | |<span class="blue-text">EXAMPLE:</span> ''CIC::DUX4'' | ||
| | |<span class="blue-text">EXAMPLE:</span> Typically, the last exon of ''CIC'' is fused to ''DUX4''. The fusion breakpoint in ''CIC'' is usually intra-exonic and removes an inhibitory sequence, upregulating ''PEA3'' genes downstream of ''CIC'' including ''ETV1'', ''ETV4'', and ''ETV5''. | ||
|<span class="blue-text">EXAMPLE:</span> t(4;19)(q25;q13) | |||
|<span class="blue-text">EXAMPLE:</span> Common (CIC-rearranged sarcoma) | |||
|<span class="blue-text">EXAMPLE:</span> D | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> | |||
''DUX4'' has many homologous genes; an alternate translocation in a minority of cases is t(10;19), but this is usually indistinguishable from t(4;19) by short-read sequencing (add references). | |||
|- | |- | ||
| | |<span class="blue-text">EXAMPLE:</span> ''ALK'' | ||
| | |<span class="blue-text">EXAMPLE:</span> ''ELM4::ALK'' | ||
< | Other fusion partners include ''KIF5B, NPM1, STRN, TFG, TPM3, CLTC, KLC1'' | ||
|<span class="blue-text">EXAMPLE:</span> Fusions result in constitutive activation of the ''ALK'' tyrosine kinase. The most common ''ALK'' fusion is ''EML4::ALK'', with breakpoints in intron 19 of ''ALK''. At the transcript level, a variable (5’) partner gene is fused to 3’ ''ALK'' at exon 20. Rarely, ''ALK'' fusions contain exon 19 due to breakpoints in intron 18. | |||
|<span class="blue-text">EXAMPLE:</span> N/A | |||
|<span class="blue-text">EXAMPLE:</span> Rare (Lung adenocarcinoma) | |||
|<span class="blue-text">EXAMPLE:</span> T | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> | |||
Both balanced and unbalanced forms are observed by FISH (add references). | |||
|- | |- | ||
|<span class="blue-text">EXAMPLE:</span> ''ABL1'' | |||
|<span class="blue-text">EXAMPLE:</span> N/A | |||
|<span class="blue-text">EXAMPLE:</span> Intragenic deletion of exons 2–7 in ''EGFR'' removes the ligand-binding domain, resulting in a constitutively active tyrosine kinase with downstream activation of multiple oncogenic pathways. | |||
|<span class="blue-text">EXAMPLE:</span> N/A | |||
|<span class="blue-text">EXAMPLE:</span> Recurrent (IDH-wildtype Glioblastoma) | |||
|<span class="blue-text">EXAMPLE:</span> D, P, T | |||
| | |||
| | |||
|- | |- | ||
| | | | ||
| | |||
| | | | ||
| | | | ||
| | | | ||
| | | | ||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Chromosomal Rearrangements (Gene Fusions)|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
NA | NA | ||
| Line 126: | Line 118: | ||
!Chromosomal Rearrangement!!Genes in Fusion (5’ or 3’ Segments)!!Pathogenic Derivative!!Prevalence | !Chromosomal Rearrangement!!Genes in Fusion (5’ or 3’ Segments)!!Pathogenic Derivative!!Prevalence | ||
|- | |- | ||
|EXAMPLE t(9;22)(q34;q11.2)||EXAMPLE 3'ABL1 / 5'BCR||EXAMPLE der(22)||EXAMPLE 5% | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 5% | ||
|- | |- | ||
|EXAMPLE t(8;21)(q22;q22)||EXAMPLE 5'RUNX1 / 3'RUNXT1||EXAMPLE der(8)||EXAMPLE 5% | |<span class="blue-text">EXAMPLE:</span> t(8;21)(q22;q22)||<span class="blue-text">EXAMPLE:</span> 5'RUNX1 / 3'RUNXT1||<span class="blue-text">EXAMPLE:</span> der(8)||<span class="blue-text">EXAMPLE:</span> 5% | ||
|} | |} | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).|Please incorporate this section into the relevant tables found in: | ||
* Chromosomal Rearrangements (Gene Fusions) | * Chromosomal Rearrangements (Gene Fusions) | ||
* Individual Region Genomic Gain/Loss/LOH | * Individual Region Genomic Gain/Loss/LOH | ||
* Characteristic Chromosomal Patterns | * Characteristic Chromosomal Patterns | ||
* Gene Mutations (SNV/INDEL)}} | * Gene Mutations (SNV/INDEL)}}</blockquote> | ||
<u>Diagnosis:</u> | <u>Diagnosis:</u> | ||
| Line 158: | Line 153: | ||
Lenalidomide is an analogue of thalidomide which functions as an immunomodulatory agent. Treatment with lenalidomide has shown great potential in reducing the abnormal clone as well as transfusion dependency irrespective of cytogenetic complexity <ref>{{Cite journal|last=Giagounidis|first=A. A. N.|last2=Germing|first2=U.|last3=Strupp|first3=C.|last4=Hildebrandt|first4=B.|last5=Heinsch|first5=M.|last6=Aul|first6=C.|date=2005|title=Prognosis of patients with del(5q) MDS and complex karyotype and the possible role of lenalidomide in this patient subgroup|url=http://link.springer.com/10.1007/s00277-005-1054-0|journal=Annals of Hematology|language=en|volume=84|issue=9|pages=569–571|doi=10.1007/s00277-005-1054-0|issn=0939-5555}}</ref><ref>{{Cite journal|last=List|first=Alan|last2=Dewald|first2=Gordon|last3=Bennett|first3=John|last4=Giagounidis|first4=Aristotle|last5=Raza|first5=Azra|last6=Feldman|first6=Eric|last7=Powell|first7=Bayard|last8=Greenberg|first8=Peter|last9=Thomas|first9=Deborah|date=2006|title=Lenalidomide in the Myelodysplastic Syndrome with Chromosome 5q Deletion|url=http://www.nejm.org/doi/abs/10.1056/NEJMoa061292|journal=New England Journal of Medicine|language=en|volume=355|issue=14|pages=1456–1465|doi=10.1056/NEJMoa061292|issn=0028-4793}}</ref>. FDA approved [https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/021880s028lbl.pdf Lenalidomide] on December 2005 for treating MDS with del(5q) with or without additional cytogenetic abnormalities. | Lenalidomide is an analogue of thalidomide which functions as an immunomodulatory agent. Treatment with lenalidomide has shown great potential in reducing the abnormal clone as well as transfusion dependency irrespective of cytogenetic complexity <ref>{{Cite journal|last=Giagounidis|first=A. A. N.|last2=Germing|first2=U.|last3=Strupp|first3=C.|last4=Hildebrandt|first4=B.|last5=Heinsch|first5=M.|last6=Aul|first6=C.|date=2005|title=Prognosis of patients with del(5q) MDS and complex karyotype and the possible role of lenalidomide in this patient subgroup|url=http://link.springer.com/10.1007/s00277-005-1054-0|journal=Annals of Hematology|language=en|volume=84|issue=9|pages=569–571|doi=10.1007/s00277-005-1054-0|issn=0939-5555}}</ref><ref>{{Cite journal|last=List|first=Alan|last2=Dewald|first2=Gordon|last3=Bennett|first3=John|last4=Giagounidis|first4=Aristotle|last5=Raza|first5=Azra|last6=Feldman|first6=Eric|last7=Powell|first7=Bayard|last8=Greenberg|first8=Peter|last9=Thomas|first9=Deborah|date=2006|title=Lenalidomide in the Myelodysplastic Syndrome with Chromosome 5q Deletion|url=http://www.nejm.org/doi/abs/10.1056/NEJMoa061292|journal=New England Journal of Medicine|language=en|volume=355|issue=14|pages=1456–1465|doi=10.1056/NEJMoa061292|issn=0028-4793}}</ref>. FDA approved [https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/021880s028lbl.pdf Lenalidomide] on December 2005 for treating MDS with del(5q) with or without additional cytogenetic abnormalities. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain/Loss/LOH== | ||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene rearrangements. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Can refer to CGC workgroup tables as linked on the homepage if applicable. Please include references throughout the table. Do not delete the table.'') </span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Chr #!!Gain | !Chr #!!Gain, Loss, Amp, LOH!!Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size]!!Relevant Gene(s) | ||
!Diagnostic | !Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | ||
!Established Clinical Significance Per Guidelines - Yes or No (Source) | |||
! | !Clinical Relevance Details/Other Notes | ||
!Notes | |||
|- | |- | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
7 | 7 | ||
|EXAMPLE Loss | |<span class="blue-text">EXAMPLE:</span> Loss | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
chr7 | chr7 | ||
| | |<span class="blue-text">EXAMPLE:</span> | ||
| | Unknown | ||
|No | |<span class="blue-text">EXAMPLE:</span> D, P | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> No | ||
|<span class="blue-text">EXAMPLE:</span> | |||
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add references). | ||
|- | |- | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
8 | 8 | ||
|EXAMPLE Gain | |<span class="blue-text">EXAMPLE:</span> Gain | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
chr8 | chr8 | ||
| | |<span class="blue-text">EXAMPLE:</span> | ||
| | Unknown | ||
| | |<span class="blue-text">EXAMPLE:</span> D, P | ||
|EXAMPLE | | | ||
|<span class="blue-text">EXAMPLE:</span> | |||
Common recurrent secondary finding for t(8;21) (add | Common recurrent secondary finding for t(8;21) (add references). | ||
|- | |||
|<span class="blue-text">EXAMPLE:</span> | |||
17 | |||
|<span class="blue-text">EXAMPLE:</span> Amp | |||
|<span class="blue-text">EXAMPLE:</span> | |||
17q12; chr17:39,700,064-39,728,658 [hg38; 28.6 kb] | |||
|<span class="blue-text">EXAMPLE:</span> | |||
''ERBB2'' | |||
|<span class="blue-text">EXAMPLE:</span> D, P, T | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> | |||
Amplification of ''ERBB2'' is associated with HER2 overexpression in HER2 positive breast cancer (add references). Add criteria for how amplification is defined. | |||
|- | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Genomic Gain/Loss/LOH|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
| Line 212: | Line 219: | ||
!Chromosome Number!!Gain/Loss/Amp/LOH!!Region | !Chromosome Number!!Gain/Loss/Amp/LOH!!Region | ||
|- | |- | ||
|EXAMPLE 8||EXAMPLE Gain||EXAMPLE chr8:0-1000000 | |<span class="blue-text">EXAMPLE:</span> 8||<span class="blue-text">EXAMPLE:</span> Gain||<span class="blue-text">EXAMPLE:</span> chr8:0-1000000 | ||
|- | |- | ||
|EXAMPLE 7||EXAMPLE Loss||EXAMPLE chr7:0-1000000 | |<span class="blue-text">EXAMPLE:</span> 7||<span class="blue-text">EXAMPLE:</span> Loss||<span class="blue-text">EXAMPLE:</span> chr7:0-1000000 | ||
|} | |} | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal or Other Global Mutational Patterns== | ||
Put your text here and fill in the table <span style="color:#0070C0">(I''nstructions: Included in this category are alterations such as hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis; microsatellite instability; homologous recombination deficiency; mutational signature pattern; etc. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.'')</span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Chromosomal Pattern | !Chromosomal Pattern | ||
! | !Molecular Pathogenesis | ||
!Prognostic Significance | !Prevalence - | ||
! | Common >20%, Recurrent 5-20% or Rare <5% (Disease) | ||
!Notes | !Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | ||
!Established Clinical Significance Per Guidelines - Yes or No (Source) | |||
!Clinical Relevance Details/Other Notes | |||
|- | |- | ||
|EXAMPLE | |<span class="blue-text">EXAMPLE:</span> | ||
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| | |<span class="blue-text">EXAMPLE:</span> See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
|<span class="blue-text">EXAMPLE:</span> Common (Oligodendroglioma) | |||
|<span class="blue-text">EXAMPLE:</span> D, P | |||
| | |||
| | |||
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | |- | ||
|<span class="blue-text">EXAMPLE:</span> | |||
Microsatellite instability - hypermutated | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> Common (Endometrial carcinoma) | |||
|<span class="blue-text">EXAMPLE:</span> P, T | |||
| | |||
| | |||
|- | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Characteristic Chromosomal Aberrations / Patterns|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
The most characteristic cytogenetic abnormality is an interstitial deletion on the long arm of chromosome 5 or del(5q)<ref name=":0">{{Cite journal|last=Van Den Berghe|first=Herman|last2=Cassiman|first2=Jean-Jacques|last3=David|first3=Guido|last4=Fryns|first4=Jean-Pierre|last5=Michaux|first5=Jean-Louis|last6=Sokal|first6=Gerard|date=1974|title=Distinct haematological disorder with deletion of long arm of No. 5 chromosome|url=http://www.nature.com/articles/251437a0|journal=Nature|language=en|volume=251|issue=5474|pages=437–438|doi=10.1038/251437a0|issn=0028-0836}}</ref> The break point is not fixed but the region between bands q31 and q33 is generally deleted. Several candidates genes were thought to contribute to the haploinsufficiency effect of the deleted region, including ''RPS14'' <ref>{{Cite journal|last=Ebert|first=Benjamin L.|last2=Pretz|first2=Jennifer|last3=Bosco|first3=Jocelyn|last4=Chang|first4=Cindy Y.|last5=Tamayo|first5=Pablo|last6=Galili|first6=Naomi|last7=Raza|first7=Azra|last8=Root|first8=David E.|last9=Attar|first9=Eyal|date=2008|title=Identification of RPS14 as a 5q- syndrome gene by RNA interference screen|url=http://www.nature.com/articles/nature06494|journal=Nature|language=en|volume=451|issue=7176|pages=335–339|doi=10.1038/nature06494|issn=0028-0836|pmc=PMC3771855|pmid=18202658}}</ref>'', CSNK1A1'' <ref name=":1">{{Cite journal|last=Schneider|first=Rebekka K.|last2=Ademà|first2=Vera|last3=Heckl|first3=Dirk|last4=Järås|first4=Marcus|last5=Mallo|first5=Mar|last6=Lord|first6=Allegra M.|last7=Chu|first7=Lisa P.|last8=McConkey|first8=Marie E.|last9=Kramann|first9=Rafael|date=2014|title=Role of Casein Kinase 1A1 in the Biology and Targeted Therapy of del(5q) MDS|url=https://linkinghub.elsevier.com/retrieve/pii/S1535610814003353|journal=Cancer Cell|language=en|volume=26|issue=4|pages=509–520|doi=10.1016/j.ccr.2014.08.001|pmc=PMC4199102|pmid=25242043}}</ref>'','' miR-145 and miR-146a <ref>{{Cite journal|last=Starczynowski|first=Daniel T|last2=Kuchenbauer|first2=Florian|last3=Argiropoulos|first3=Bob|last4=Sung|first4=Sandy|last5=Morin|first5=Ryan|last6=Muranyi|first6=Andrew|last7=Hirst|first7=Martin|last8=Hogge|first8=Donna|last9=Marra|first9=Marco|date=2010|title=Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype|url=http://www.nature.com/articles/nm.2054|journal=Nature Medicine|language=en|volume=16|issue=1|pages=49–58|doi=10.1038/nm.2054|issn=1078-8956}}</ref>. Identification of del(5q) has clinical significance because patients with this cytogenetic abnormality have a good prognosis and they respond well to lenalidomide treatment. | The most characteristic cytogenetic abnormality is an interstitial deletion on the long arm of chromosome 5 or del(5q)<ref name=":0">{{Cite journal|last=Van Den Berghe|first=Herman|last2=Cassiman|first2=Jean-Jacques|last3=David|first3=Guido|last4=Fryns|first4=Jean-Pierre|last5=Michaux|first5=Jean-Louis|last6=Sokal|first6=Gerard|date=1974|title=Distinct haematological disorder with deletion of long arm of No. 5 chromosome|url=http://www.nature.com/articles/251437a0|journal=Nature|language=en|volume=251|issue=5474|pages=437–438|doi=10.1038/251437a0|issn=0028-0836}}</ref> The break point is not fixed but the region between bands q31 and q33 is generally deleted. Several candidates genes were thought to contribute to the haploinsufficiency effect of the deleted region, including ''RPS14'' <ref>{{Cite journal|last=Ebert|first=Benjamin L.|last2=Pretz|first2=Jennifer|last3=Bosco|first3=Jocelyn|last4=Chang|first4=Cindy Y.|last5=Tamayo|first5=Pablo|last6=Galili|first6=Naomi|last7=Raza|first7=Azra|last8=Root|first8=David E.|last9=Attar|first9=Eyal|date=2008|title=Identification of RPS14 as a 5q- syndrome gene by RNA interference screen|url=http://www.nature.com/articles/nature06494|journal=Nature|language=en|volume=451|issue=7176|pages=335–339|doi=10.1038/nature06494|issn=0028-0836|pmc=PMC3771855|pmid=18202658}}</ref>'', CSNK1A1'' <ref name=":1">{{Cite journal|last=Schneider|first=Rebekka K.|last2=Ademà|first2=Vera|last3=Heckl|first3=Dirk|last4=Järås|first4=Marcus|last5=Mallo|first5=Mar|last6=Lord|first6=Allegra M.|last7=Chu|first7=Lisa P.|last8=McConkey|first8=Marie E.|last9=Kramann|first9=Rafael|date=2014|title=Role of Casein Kinase 1A1 in the Biology and Targeted Therapy of del(5q) MDS|url=https://linkinghub.elsevier.com/retrieve/pii/S1535610814003353|journal=Cancer Cell|language=en|volume=26|issue=4|pages=509–520|doi=10.1016/j.ccr.2014.08.001|pmc=PMC4199102|pmid=25242043}}</ref>'','' miR-145 and miR-146a <ref>{{Cite journal|last=Starczynowski|first=Daniel T|last2=Kuchenbauer|first2=Florian|last3=Argiropoulos|first3=Bob|last4=Sung|first4=Sandy|last5=Morin|first5=Ryan|last6=Muranyi|first6=Andrew|last7=Hirst|first7=Martin|last8=Hogge|first8=Donna|last9=Marra|first9=Marco|date=2010|title=Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype|url=http://www.nature.com/articles/nm.2054|journal=Nature Medicine|language=en|volume=16|issue=1|pages=49–58|doi=10.1038/nm.2054|issn=1078-8956}}</ref>. Identification of del(5q) has clinical significance because patients with this cytogenetic abnormality have a good prognosis and they respond well to lenalidomide treatment. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV/INDEL)== | ||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent or common as well either disease defining and/or clinically significant. If a gene has multiple mechanisms depending on the type or site of the alteration, add multiple entries in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Details on clinical significance such as prognosis and other important information such as concomitant and mutually exclusive mutations can be provided in the notes section. Please include references throughout the table. Do not delete the table.'') </span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene | !Gene!!Genetic Alteration!!Tumor Suppressor Gene, Oncogene, Other!!Prevalence - | ||
! | Common >20%, Recurrent 5-20% or Rare <5% (Disease) | ||
!Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | |||
! | !Established Clinical Significance Per Guidelines - Yes or No (Source) | ||
!Notes | !Clinical Relevance Details/Other Notes | ||
|- | |- | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span>''EGFR'' | ||
EXAMPLE: | <br /> | ||
|<span class="blue-text">EXAMPLE:</span> Exon 18-21 activating mutations | |||
|<span class="blue-text">EXAMPLE:</span> Oncogene | |||
|<span class="blue-text">EXAMPLE:</span> Common (lung cancer) | |||
EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> T | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Yes (NCCN) | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Exons 18, 19, and 21 mutations are targetable for therapy. Exon 20 T790M variants cause resistance to first generation TKI therapy and are targetable by second and third generation TKIs (add references). | ||
|- | |||
EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> ''TP53''; Variable LOF mutations | ||
|EXAMPLE: | <br /> | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Variable LOF mutations | ||
|<span class="blue-text">EXAMPLE:</span> Tumor Supressor Gene | |||
|<span class="blue-text">EXAMPLE:</span> Common (breast cancer) | |||
|<span class="blue-text">EXAMPLE:</span> P | |||
| | |||
|<span class="blue-text">EXAMPLE:</span> >90% are somatic; rare germline alterations associated with Li-Fraumeni syndrome (add reference). Denotes a poor prognosis in breast cancer. | |||
|- | |||
|<span class="blue-text">EXAMPLE:</span> ''BRAF''; Activating mutations | |||
|<span class="blue-text">EXAMPLE:</span> Activating mutations | |||
|<span class="blue-text">EXAMPLE:</span> Oncogene | |||
|<span class="blue-text">EXAMPLE:</span> Common (melanoma) | |||
|<span class="blue-text">EXAMPLE:</span> T | |||
| | |||
| | |||
|- | |||
| | |||
| | |||
| | |||
| | |||
| | | | ||
| | | | ||
| | | | ||
|}Note: A more extensive list of mutations can be found in [https://www.cbioportal.org/ <u>cBioportal</u>], [https://cancer.sanger.ac.uk/cosmic <u>COSMIC</u>], and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | |||
|} | |||
Note: A more extensive list of mutations can be found in | |||
<blockquote class="blockedit">{{Box-round|title=v4:Gene Mutations (SNV/INDEL)|The content below was from the old template. Please incorporate above.}}</blockquote> | |||
<blockquote class= | |||
Somatic mutations in ''JAK2'' and ''MPL'' have been reported in a small subset patients with isolated del(5q), but these mutations seem not confer diagnostic or prognostic value <ref>{{Cite journal|last=Patnaik|first=M M|last2=Lasho|first2=T L|last3=Finke|first3=C M|last4=Gangat|first4=N|last5=Caramazza|first5=D|last6=Holtan|first6=S G|last7=Pardanani|first7=A|last8=Knudson|first8=R A|last9=Ketterling|first9=R P|date=2010|title=WHO-defined ‘myelodysplastic syndrome with isolated del(5q)’ in 88 consecutive patients: survival data, leukemic transformation rates and prevalence of JAK2, MPL and IDH mutations|url=http://www.nature.com/articles/leu2010105|journal=Leukemia|language=en|volume=24|issue=7|pages=1283–1289|doi=10.1038/leu.2010.105|issn=0887-6924|pmc=PMC3035970|pmid=20485371}}</ref>. A subset of cases could have ''[[SF3B1]]'' mutations, which needs to be differentiated with MDS-RS. | Somatic mutations in ''JAK2'' and ''MPL'' have been reported in a small subset patients with isolated del(5q), but these mutations seem not confer diagnostic or prognostic value <ref>{{Cite journal|last=Patnaik|first=M M|last2=Lasho|first2=T L|last3=Finke|first3=C M|last4=Gangat|first4=N|last5=Caramazza|first5=D|last6=Holtan|first6=S G|last7=Pardanani|first7=A|last8=Knudson|first8=R A|last9=Ketterling|first9=R P|date=2010|title=WHO-defined ‘myelodysplastic syndrome with isolated del(5q)’ in 88 consecutive patients: survival data, leukemic transformation rates and prevalence of JAK2, MPL and IDH mutations|url=http://www.nature.com/articles/leu2010105|journal=Leukemia|language=en|volume=24|issue=7|pages=1283–1289|doi=10.1038/leu.2010.105|issn=0887-6924|pmc=PMC3035970|pmid=20485371}}</ref>. A subset of cases could have ''[[SF3B1]]'' mutations, which needs to be differentiated with MDS-RS. | ||
| Line 287: | Line 329: | ||
!Gene!!Mutation!!Oncogene/Tumor Suppressor/Other!!Presumed Mechanism (LOF/GOF/Other; Driver/Passenger)!!Prevalence (COSMIC/TCGA/Other) | !Gene!!Mutation!!Oncogene/Tumor Suppressor/Other!!Presumed Mechanism (LOF/GOF/Other; Driver/Passenger)!!Prevalence (COSMIC/TCGA/Other) | ||
|- | |- | ||
|EXAMPLE TP53||EXAMPLE R273H||EXAMPLE Tumor Suppressor||EXAMPLE LOF||EXAMPLE 20% | |<span class="blue-text">EXAMPLE:</span> TP53||<span class="blue-text">EXAMPLE:</span> R273H||<span class="blue-text">EXAMPLE:</span> Tumor Suppressor||<span class="blue-text">EXAMPLE:</span> LOF||<span class="blue-text">EXAMPLE:</span> 20% | ||
|} | |} | ||
| Line 302: | Line 344: | ||
|} | |} | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Epigenomic Alterations== | ==Epigenomic Alterations== | ||
| Line 309: | Line 354: | ||
==Genes and Main Pathways Involved== | ==Genes and Main Pathways Involved== | ||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: | |||
Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Please include references throughout the table. Do not delete the table.)''</span> | |||
{| class="wikitable sortable" | {| class="wikitable sortable" | ||
|- | |- | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
|EXAMPLE: BRAF and MAP2K1; Activating mutations | |<span class="blue-text">EXAMPLE:</span> ''BRAF'' and ''MAP2K1''; Activating mutations | ||
|EXAMPLE: MAPK signaling | |<span class="blue-text">EXAMPLE:</span> MAPK signaling | ||
|EXAMPLE: Increased cell growth and proliferation | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation | ||
|- | |- | ||
|EXAMPLE: CDKN2A; Inactivating mutations | |<span class="blue-text">EXAMPLE:</span> ''CDKN2A''; Inactivating mutations | ||
|EXAMPLE: Cell cycle regulation | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation | ||
|EXAMPLE: Unregulated cell division | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division | ||
|- | |- | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> ''KMT2C'' and ''ARID1A''; Inactivating mutations | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling | ||
|EXAMPLE: | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program | ||
|- | |||
| | |||
| | |||
| | |||
|} | |} | ||
<blockquote class= | <blockquote class="blockedit">{{Box-round|title=v4:Genes and Main Pathways Involved|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
Several candidate genes in the common deleted region of 5q have been reported to contribute to the molecular pathogenesis. | Several candidate genes in the common deleted region of 5q have been reported to contribute to the molecular pathogenesis. | ||
| Line 337: | Line 387: | ||
miR-145 and miR146α: These two micro RNA genes are thought to be associated with thrombocytosis and hypolobulated megakaryocytes <ref>{{Cite journal|last=Starczynowski|first=Daniel T|last2=Kuchenbauer|first2=Florian|last3=Argiropoulos|first3=Bob|last4=Sung|first4=Sandy|last5=Morin|first5=Ryan|last6=Muranyi|first6=Andrew|last7=Hirst|first7=Martin|last8=Hogge|first8=Donna|last9=Marra|first9=Marco|date=2010|title=Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype|url=http://www.nature.com/articles/nm.2054|journal=Nature Medicine|language=en|volume=16|issue=1|pages=49–58|doi=10.1038/nm.2054|issn=1078-8956}}</ref>. | miR-145 and miR146α: These two micro RNA genes are thought to be associated with thrombocytosis and hypolobulated megakaryocytes <ref>{{Cite journal|last=Starczynowski|first=Daniel T|last2=Kuchenbauer|first2=Florian|last3=Argiropoulos|first3=Bob|last4=Sung|first4=Sandy|last5=Morin|first5=Ryan|last6=Muranyi|first6=Andrew|last7=Hirst|first7=Martin|last8=Hogge|first8=Donna|last9=Marra|first9=Marco|date=2010|title=Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype|url=http://www.nature.com/articles/nm.2054|journal=Nature Medicine|language=en|volume=16|issue=1|pages=49–58|doi=10.1038/nm.2054|issn=1078-8956}}</ref>. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Genetic Diagnostic Testing Methods== | ==Genetic Diagnostic Testing Methods== | ||
| Line 345: | Line 398: | ||
==Familial Forms== | ==Familial Forms== | ||
Put your text here <span style="color:#0070C0">(''Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.'') </span> | Put your text here <span style="color:#0070C0">(''Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.'') </span> | ||
==Additional Information== | ==Additional Information== | ||
| Line 357: | Line 410: | ||
==References== | ==References== | ||
(use the "Cite" icon at the top of the page) <span style="color:#0070C0">(''Instructions: Add each reference into the text above by clicking | (use the "Cite" icon at the top of the page) <span style="color:#0070C0">(''Instructions: Add each reference into the text above by clicking where you want to insert the reference, selecting the “Cite” icon at the top of the wiki page, and using the “Automatic” tab option to search by PMID to select the reference to insert. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference. To insert the same reference again later in the page, select the “Cite” icon and “Re-use” to find the reference; DO NOT insert the same reference twice using the “Automatic” tab as it will be treated as two separate references. The reference list in this section will be automatically generated and sorted''</span><span style="color:#0070C0">''.''</span><span style="color:#0070C0">)</span> <references /> | ||
<br /> | |||
==Notes== | ==Notes== | ||
<nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the [[Leadership|''<u>Associate Editor</u>'']] or other CCGA representative. When pages have a major update, the new author will be acknowledged at the beginning of the page, and those who contributed previously will be acknowledged below as a prior author. | ||
Prior Author(s): | |||
<nowiki>*</nowiki>''Citation of this Page'': “Myelodysplastic neoplasm with low blasts and 5q deletion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/HAEM5:Myelodysplastic_neoplasm_with_low_blasts_and_5q_deletion</nowiki>. | <nowiki>*</nowiki>''Citation of this Page'': “Myelodysplastic neoplasm with low blasts and 5q deletion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/HAEM5:Myelodysplastic_neoplasm_with_low_blasts_and_5q_deletion</nowiki>. | ||
[[Category:HAEM5]][[Category:DISEASE]][[Category:Diseases M]] | [[Category:HAEM5]] | ||
[[Category:DISEASE]] | |||
[[Category:Diseases M]] | |||
Latest revision as of 12:22, 3 July 2025
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Myelodysplastic Syndrome (MDS) with Isolated del(5q).
(General Instructions – The focus of these pages is the clinically significant genetic alterations in each disease type. This is based on up-to-date knowledge from multiple resources such as PubMed and the WHO classification books. The CCGA is meant to be a supplemental resource to the WHO classification books; the CCGA captures in a continually updated wiki-stye manner the current genetics/genomics knowledge of each disease, which evolves more rapidly than books can be revised and published. If the same disease is described in multiple WHO classification books, the genetics-related information for that disease will be consolidated into a single main page that has this template (other pages would only contain a link to this main page). Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column in a table, click nearby within the table and select the > symbol that appears. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support.)
Primary Author(s)*
Xiaolin Hu, Ph.D; Teresa Smolarek, Ph.D, FACMG
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Myelodysplastic neoplasms |
| Type | Myelodysplastic neoplasms, with defining genetic abnormalities |
| Subtype(s) | Myelodysplastic neoplasm with low blasts and 5q deletion |
Related Terminology
| Acceptable | Myelodysplastic syndrome with del(5q) |
| Not Recommended | 5q− syndrome |
Gene Rearrangements
Put your text here and fill in the table (Instructions: Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Driver Gene | Fusion(s) and Common Partner Genes | Molecular Pathogenesis | Typical Chromosomal Alteration(s) | Prevalence -Common >20%, Recurrent 5-20% or Rare <5% (Disease) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: ABL1 | EXAMPLE: BCR::ABL1 | EXAMPLE: The pathogenic derivative is the der(22) resulting in fusion of 5’ BCR and 3’ABL1. | EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: Common (CML) | EXAMPLE: D, P, T | EXAMPLE: Yes (WHO, NCCN) | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). BCR::ABL1 is generally favorable in CML (add reference). |
| EXAMPLE: CIC | EXAMPLE: CIC::DUX4 | EXAMPLE: Typically, the last exon of CIC is fused to DUX4. The fusion breakpoint in CIC is usually intra-exonic and removes an inhibitory sequence, upregulating PEA3 genes downstream of CIC including ETV1, ETV4, and ETV5. | EXAMPLE: t(4;19)(q25;q13) | EXAMPLE: Common (CIC-rearranged sarcoma) | EXAMPLE: D | EXAMPLE:
DUX4 has many homologous genes; an alternate translocation in a minority of cases is t(10;19), but this is usually indistinguishable from t(4;19) by short-read sequencing (add references). | |
| EXAMPLE: ALK | EXAMPLE: ELM4::ALK
|
EXAMPLE: Fusions result in constitutive activation of the ALK tyrosine kinase. The most common ALK fusion is EML4::ALK, with breakpoints in intron 19 of ALK. At the transcript level, a variable (5’) partner gene is fused to 3’ ALK at exon 20. Rarely, ALK fusions contain exon 19 due to breakpoints in intron 18. | EXAMPLE: N/A | EXAMPLE: Rare (Lung adenocarcinoma) | EXAMPLE: T | EXAMPLE:
Both balanced and unbalanced forms are observed by FISH (add references). | |
| EXAMPLE: ABL1 | EXAMPLE: N/A | EXAMPLE: Intragenic deletion of exons 2–7 in EGFR removes the ligand-binding domain, resulting in a constitutively active tyrosine kinase with downstream activation of multiple oncogenic pathways. | EXAMPLE: N/A | EXAMPLE: Recurrent (IDH-wildtype Glioblastoma) | EXAMPLE: D, P, T | ||
editv4:Chromosomal Rearrangements (Gene Fusions)The content below was from the old template. Please incorporate above.
NA
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence |
|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 5% |
| EXAMPLE: t(8;21)(q22;q22) | EXAMPLE: 5'RUNX1 / 3'RUNXT1 | EXAMPLE: der(8) | EXAMPLE: 5% |
End of V4 Section
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
Diagnosis:
- Cytopenia in 1 to 2 lineages. Cytopenias are defined as hemoglobin concentration < 10 g/dL, platelet count < 100 x 109/L and absolute neutrophil count < 1.8 x 109/L; mild degree of anemia (hemoglobin < 13 g/dL in men or < 12 g/dL in women) or thrombocytopenia (platelets < 150 x109/L) are allowed if defining cytogenetic abnormality is present. Thrombocytosis (platelet count ≥ 450 x 109/L) is allowed in MDS with isolated del(5q). PB monocytes must be < 1 x109/L.
- Dysplasia in 1 to 3 lineages. Micromegakaryocytes with hypolobation are characteristic dysplasia. Ring sideroblasts are not common.
- Blasts percentage: BM <5%, PB <1%, no Auer rods.
- Interstitial deletion of 5q alone or with an additional abnormality other than loss of chromosome 7 or deletion of 7q.
- Chromosome 7 loss or del(7q), more than one additional cytogenetic abnormality, excess blasts need to be excluded from the diagnosis.
Prognosis:
- According to IPSS‐R good‐risk group with low rates of leukemic transformation [1].
- The median survival of MDS with isolated del(5q) is 66 to 145 months.
- About less than 10% of cases progress to acute myeloid leukemia [2].
- Mutations of TP53 may function as an independent prognostic factor in associated with leukemic progression and resistant to lenalidomide[3][4].
Therapeutic Implications:
Lenalidomide is an analogue of thalidomide which functions as an immunomodulatory agent. Treatment with lenalidomide has shown great potential in reducing the abnormal clone as well as transfusion dependency irrespective of cytogenetic complexity [5][6]. FDA approved Lenalidomide on December 2005 for treating MDS with del(5q) with or without additional cytogenetic abnormalities.
End of V4 Section
Individual Region Genomic Gain/Loss/LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene rearrangements. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Can refer to CGC workgroup tables as linked on the homepage if applicable. Please include references throughout the table. Do not delete the table.)
| Chr # | Gain, Loss, Amp, LOH | Minimal Region Cytoband and/or Genomic Coordinates [Genome Build; Size] | Relevant Gene(s) | Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7 |
EXAMPLE:
Unknown |
EXAMPLE: D, P | EXAMPLE: No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add references). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8 |
EXAMPLE:
Unknown |
EXAMPLE: D, P | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add references). | |
| EXAMPLE:
17 |
EXAMPLE: Amp | EXAMPLE:
17q12; chr17:39,700,064-39,728,658 [hg38; 28.6 kb] |
EXAMPLE:
ERBB2 |
EXAMPLE: D, P, T | EXAMPLE:
Amplification of ERBB2 is associated with HER2 overexpression in HER2 positive breast cancer (add references). Add criteria for how amplification is defined. | |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.
| Chromosome Number | Gain/Loss/Amp/LOH | Region |
|---|---|---|
| EXAMPLE: 8 | EXAMPLE: Gain | EXAMPLE: chr8:0-1000000 |
| EXAMPLE: 7 | EXAMPLE: Loss | EXAMPLE: chr7:0-1000000 |
End of V4 Section
Characteristic Chromosomal or Other Global Mutational Patterns
Put your text here and fill in the table (Instructions: Included in this category are alterations such as hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis; microsatellite instability; homologous recombination deficiency; mutational signature pattern; etc. Details on clinical significance such as prognosis and other important information can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Chromosomal Pattern | Molecular Pathogenesis | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
EXAMPLE: See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | EXAMPLE: Common (Oligodendroglioma) | EXAMPLE: D, P | ||
| EXAMPLE:
Microsatellite instability - hypermutated |
EXAMPLE: Common (Endometrial carcinoma) | EXAMPLE: P, T | |||
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.
The most characteristic cytogenetic abnormality is an interstitial deletion on the long arm of chromosome 5 or del(5q)[7] The break point is not fixed but the region between bands q31 and q33 is generally deleted. Several candidates genes were thought to contribute to the haploinsufficiency effect of the deleted region, including RPS14 [8], CSNK1A1 [9], miR-145 and miR-146a [10]. Identification of del(5q) has clinical significance because patients with this cytogenetic abnormality have a good prognosis and they respond well to lenalidomide treatment.
End of V4 Section
Gene Mutations (SNV/INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent or common as well either disease defining and/or clinically significant. If a gene has multiple mechanisms depending on the type or site of the alteration, add multiple entries in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Details on clinical significance such as prognosis and other important information such as concomitant and mutually exclusive mutations can be provided in the notes section. Please include references throughout the table. Do not delete the table.)
| Gene | Genetic Alteration | Tumor Suppressor Gene, Oncogene, Other | Prevalence -
Common >20%, Recurrent 5-20% or Rare <5% (Disease) |
Diagnostic, Prognostic, and Therapeutic Significance - D, P, T | Established Clinical Significance Per Guidelines - Yes or No (Source) | Clinical Relevance Details/Other Notes |
|---|---|---|---|---|---|---|
| EXAMPLE:EGFR
|
EXAMPLE: Exon 18-21 activating mutations | EXAMPLE: Oncogene | EXAMPLE: Common (lung cancer) | EXAMPLE: T | EXAMPLE: Yes (NCCN) | EXAMPLE: Exons 18, 19, and 21 mutations are targetable for therapy. Exon 20 T790M variants cause resistance to first generation TKI therapy and are targetable by second and third generation TKIs (add references). |
| EXAMPLE: TP53; Variable LOF mutations
|
EXAMPLE: Variable LOF mutations | EXAMPLE: Tumor Supressor Gene | EXAMPLE: Common (breast cancer) | EXAMPLE: P | EXAMPLE: >90% are somatic; rare germline alterations associated with Li-Fraumeni syndrome (add reference). Denotes a poor prognosis in breast cancer. | |
| EXAMPLE: BRAF; Activating mutations | EXAMPLE: Activating mutations | EXAMPLE: Oncogene | EXAMPLE: Common (melanoma) | EXAMPLE: T | ||
Note: A more extensive list of mutations can be found in cBioportal, COSMIC, and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.
Somatic mutations in JAK2 and MPL have been reported in a small subset patients with isolated del(5q), but these mutations seem not confer diagnostic or prognostic value [11]. A subset of cases could have SF3B1 mutations, which needs to be differentiated with MDS-RS.
| Gene | Mutation | Oncogene/Tumor Suppressor/Other | Presumed Mechanism (LOF/GOF/Other; Driver/Passenger) | Prevalence (COSMIC/TCGA/Other) |
|---|---|---|---|---|
| EXAMPLE: TP53 | EXAMPLE: R273H | EXAMPLE: Tumor Suppressor | EXAMPLE: LOF | EXAMPLE: 20% |
Other Mutations
| Type | Gene/Region/Other |
|---|---|
| Concomitant Mutations | JAK2 V617F, MPL W515L |
| Secondary Mutations | |
| Mutually Exclusive |
End of V4 Section
Epigenomic Alterations
Genes involved in epigenetic regulation are frequently mutated in MDS such as TET2, DNMT3A, IDH1, IDH2, AXSL1, and EZH2 [12]. These genes play a role in DNA methylation and chromatin modification as well as regulating gene expression.
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Please include references throughout the table. Do not delete the table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.
Several candidate genes in the common deleted region of 5q have been reported to contribute to the molecular pathogenesis.
RPS14: Encodes for ribosomal subunit of 40S. Inactivation of RPS14 results in ineffective erythroid differentiation and increased apoptosis in a p53 dependent way [13].
CSNK1A1: Encodes for CK1α, a serine/threonine protein kinase activity. Mutations in CSNK1A1 lead to clonal expansion of hematopoietic stem cells through activation of β-catenin [9].
miR-145 and miR146α: These two micro RNA genes are thought to be associated with thrombocytosis and hypolobulated megakaryocytes [14].
End of V4 Section
Genetic Diagnostic Testing Methods
- Bone marrow morphology and peripheral blood test are standard in diagnosis of MDS.
- FISH probes targeting the CDR are commonly incorporated into MDS panel to assess -5/5q-.
- Conventional cytogenetics are also helpful in diagnosis.
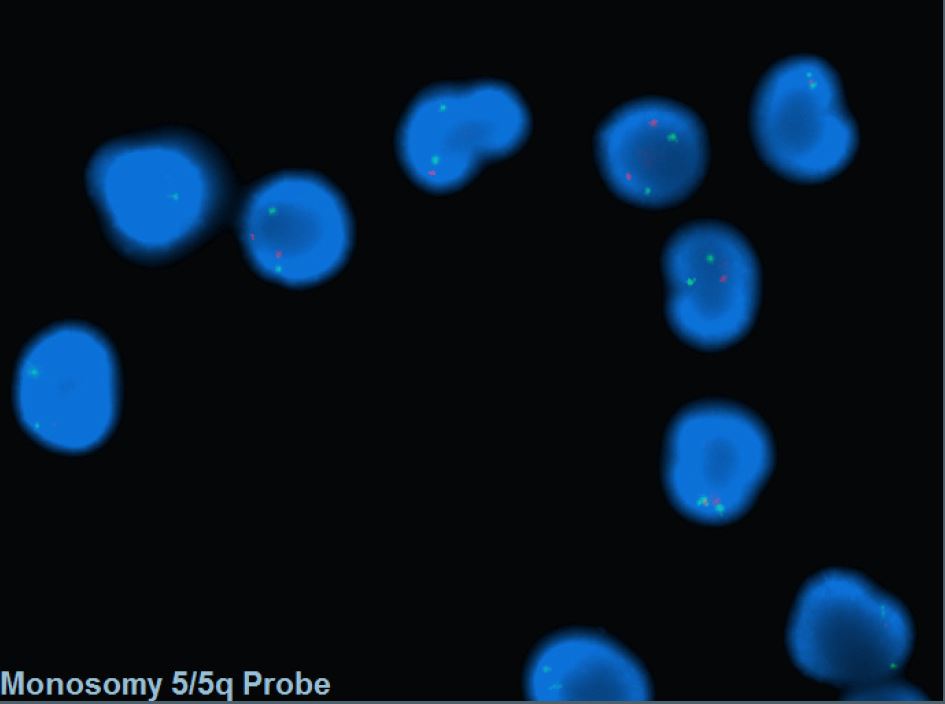
Fluorescent in situ hybridization (FISH) with a dual color DNA probe specific for the EGR1 (5q31.2) locus and to a distal short arm locus (5p15.31) of chromosome 5 detected two green and one red signal pattern that consistents with 5q deletion in 51% of patient's cells.
Familial Forms
Put your text here (Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.)
Additional Information
Put your text here
Links
Put your links here (use link icon at top of page)
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking where you want to insert the reference, selecting the “Cite” icon at the top of the wiki page, and using the “Automatic” tab option to search by PMID to select the reference to insert. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference. To insert the same reference again later in the page, select the “Cite” icon and “Re-use” to find the reference; DO NOT insert the same reference twice using the “Automatic” tab as it will be treated as two separate references. The reference list in this section will be automatically generated and sorted.)
- ↑ Mallo, M; et al. (2011). "Impact of adjunct cytogenetic abnormalities for prognostic stratification in patients with myelodysplastic syndrome and deletion 5q". Leukemia. 25 (1): 110–120. doi:10.1038/leu.2010.231. ISSN 0887-6924.
- ↑ Boultwood, Jacqueline; et al. (2010). "Advances in the 5q− syndrome". Blood. 116 (26): 5803–5811. doi:10.1182/blood-2010-04-273771. ISSN 0006-4971.
- ↑ Jädersten, Martin; et al. (2011). "TP53 Mutations in Low-Risk Myelodysplastic Syndromes With del(5q) Predict Disease Progression". Journal of Clinical Oncology. 29 (15): 1971–1979. doi:10.1200/JCO.2010.31.8576. ISSN 0732-183X.
- ↑ Kulasekararaj, Austin G.; et al. (2013). "TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis". British Journal of Haematology. 160 (5): 660–672. doi:10.1111/bjh.12203.
- ↑ Giagounidis, A. A. N.; et al. (2005). "Prognosis of patients with del(5q) MDS and complex karyotype and the possible role of lenalidomide in this patient subgroup". Annals of Hematology. 84 (9): 569–571. doi:10.1007/s00277-005-1054-0. ISSN 0939-5555.
- ↑ List, Alan; et al. (2006). "Lenalidomide in the Myelodysplastic Syndrome with Chromosome 5q Deletion". New England Journal of Medicine. 355 (14): 1456–1465. doi:10.1056/NEJMoa061292. ISSN 0028-4793.
- ↑ Van Den Berghe, Herman; et al. (1974). "Distinct haematological disorder with deletion of long arm of No. 5 chromosome". Nature. 251 (5474): 437–438. doi:10.1038/251437a0. ISSN 0028-0836.
- ↑ Ebert, Benjamin L.; et al. (2008). "Identification of RPS14 as a 5q- syndrome gene by RNA interference screen". Nature. 451 (7176): 335–339. doi:10.1038/nature06494. ISSN 0028-0836. PMC 3771855. PMID 18202658.CS1 maint: PMC format (link)
- ↑ 9.0 9.1 Schneider, Rebekka K.; et al. (2014). "Role of Casein Kinase 1A1 in the Biology and Targeted Therapy of del(5q) MDS". Cancer Cell. 26 (4): 509–520. doi:10.1016/j.ccr.2014.08.001. PMC 4199102. PMID 25242043.CS1 maint: PMC format (link)
- ↑ Starczynowski, Daniel T; et al. (2010). "Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype". Nature Medicine. 16 (1): 49–58. doi:10.1038/nm.2054. ISSN 1078-8956.
- ↑ Patnaik, M M; et al. (2010). "WHO-defined 'myelodysplastic syndrome with isolated del(5q)' in 88 consecutive patients: survival data, leukemic transformation rates and prevalence of JAK2, MPL and IDH mutations". Leukemia. 24 (7): 1283–1289. doi:10.1038/leu.2010.105. ISSN 0887-6924. PMC 3035970. PMID 20485371.CS1 maint: PMC format (link)
- ↑ Heuser, Michael; et al. (2018). "Epigenetics in myelodysplastic syndromes". Seminars in Cancer Biology. 51: 170–179. doi:10.1016/j.semcancer.2017.07.009.
- ↑ Barlow, Jillian L; et al. (2010). "A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q– syndrome". Nature Medicine. 16 (1): 59–66. doi:10.1038/nm.2063. ISSN 1078-8956. PMC 2803774. PMID 19966810.CS1 maint: PMC format (link)
- ↑ Starczynowski, Daniel T; et al. (2010). "Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype". Nature Medicine. 16 (1): 49–58. doi:10.1038/nm.2054. ISSN 1078-8956.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the Associate Editor or other CCGA representative. When pages have a major update, the new author will be acknowledged at the beginning of the page, and those who contributed previously will be acknowledged below as a prior author.
Prior Author(s):
*Citation of this Page: “Myelodysplastic neoplasm with low blasts and 5q deletion”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 07/3/2025, https://ccga.io/index.php/HAEM5:Myelodysplastic_neoplasm_with_low_blasts_and_5q_deletion.