HAEM5:Acute myeloid leukaemia with MECOM rearrangement: Difference between revisions
| [checked revision] | [checked revision] |
Bailey.Glen (talk | contribs) No edit summary |
Bailey.Glen (talk | contribs) No edit summary |
||
| Line 82: | Line 82: | ||
<blockquote class='blockedit'>{{Box-round|title=v4:Clinical Features|The content below was from the old template. Please incorporate above.}} | <blockquote class='blockedit'>{{Box-round|title=v4:Clinical Features|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
The patients often present with anemia and normal or increased platelet counts. However, decreased platelet counts and hepatosplenomegaly can also be observed<ref name=":0" /><ref name=":1" />. | The patients often present with anemia and normal or increased platelet counts. However, decreased platelet counts and hepatosplenomegaly can also be observed<ref name=":0" /><ref name=":1" />. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Sites of Involvement== | ==Sites of Involvement== | ||
| Line 140: | Line 143: | ||
* Individual Region Genomic Gain/Loss/LOH | * Individual Region Genomic Gain/Loss/LOH | ||
* Characteristic Chromosomal Patterns | * Characteristic Chromosomal Patterns | ||
* Gene Mutations (SNV/INDEL)}} | * Gene Mutations (SNV/INDEL)}}</blockquote> | ||
Diagnostic: The presence of inv(3)/t(3;3) defines a cytogenetic subtype of AML, however in the current WHO 2017 classification does not allow to make a diagnosis of AML if blast percentage <20%. Very poor prognosis and rapid progression of MDS with inv(3)(q21q26.2)/t(3;3)(q21;q26.2) resulted in a proposal to consider neoplasms with these rearrangements as an AML with recurrent genetic abnormalities, irrespective of blast percentage. | Diagnostic: The presence of inv(3)/t(3;3) defines a cytogenetic subtype of AML, however in the current WHO 2017 classification does not allow to make a diagnosis of AML if blast percentage <20%. Very poor prognosis and rapid progression of MDS with inv(3)(q21q26.2)/t(3;3)(q21;q26.2) resulted in a proposal to consider neoplasms with these rearrangements as an AML with recurrent genetic abnormalities, irrespective of blast percentage. | ||
| Line 150: | Line 153: | ||
Therapeutic: AML with inv(3)(q21q26.2) or t(3;3) (q21;q26.2) is an aggressive disease with short survival, in need for novel and more efficient treatments. Some studies have shown that arsenic trioxide induces targeted specific degradation of the AML1/MDS1/EVI1 oncoprotein, and an isolated case report described good response to Arsenic trioxide and thalidomide combination in a patient with MDS with inv(3)<ref>{{Cite journal|last=Shackelford|first=David|last2=Kenific|first2=Candia|last3=Blusztajn|first3=Agnieszka|last4=Waxman|first4=Samuel|last5=Ren|first5=Ruibao|date=2006|title=Targeted degradation of the AML1/MDS1/EVI1 oncoprotein by arsenic trioxide|url=https://www.ncbi.nlm.nih.gov/pubmed/17145882|journal=Cancer Research|volume=66|issue=23|pages=11360–11369|doi=10.1158/0008-5472.CAN-06-1774|issn=0008-5472|pmid=17145882}}</ref><ref>{{Cite journal|last=Raza|first=Azra|last2=Buonamici|first2=Silvia|last3=Lisak|first3=Laurie|last4=Tahir|first4=Sarah|last5=Li|first5=Donglan|last6=Imran|first6=Mehnaz|last7=Chaudary|first7=Nusrat Ijaz|last8=Pervaiz|first8=Hassan|last9=Gallegos|first9=J. Alejandro|date=2004|title=Arsenic trioxide and thalidomide combination produces multi-lineage hematological responses in myelodysplastic syndromes patients, particularly in those with high pre-therapy EVI1 expression|url=https://www.ncbi.nlm.nih.gov/pubmed/15203277|journal=Leukemia Research|volume=28|issue=8|pages=791–803|doi=10.1016/j.leukres.2003.11.018|issn=0145-2126|pmid=15203277}}</ref>. | Therapeutic: AML with inv(3)(q21q26.2) or t(3;3) (q21;q26.2) is an aggressive disease with short survival, in need for novel and more efficient treatments. Some studies have shown that arsenic trioxide induces targeted specific degradation of the AML1/MDS1/EVI1 oncoprotein, and an isolated case report described good response to Arsenic trioxide and thalidomide combination in a patient with MDS with inv(3)<ref>{{Cite journal|last=Shackelford|first=David|last2=Kenific|first2=Candia|last3=Blusztajn|first3=Agnieszka|last4=Waxman|first4=Samuel|last5=Ren|first5=Ruibao|date=2006|title=Targeted degradation of the AML1/MDS1/EVI1 oncoprotein by arsenic trioxide|url=https://www.ncbi.nlm.nih.gov/pubmed/17145882|journal=Cancer Research|volume=66|issue=23|pages=11360–11369|doi=10.1158/0008-5472.CAN-06-1774|issn=0008-5472|pmid=17145882}}</ref><ref>{{Cite journal|last=Raza|first=Azra|last2=Buonamici|first2=Silvia|last3=Lisak|first3=Laurie|last4=Tahir|first4=Sarah|last5=Li|first5=Donglan|last6=Imran|first6=Mehnaz|last7=Chaudary|first7=Nusrat Ijaz|last8=Pervaiz|first8=Hassan|last9=Gallegos|first9=J. Alejandro|date=2004|title=Arsenic trioxide and thalidomide combination produces multi-lineage hematological responses in myelodysplastic syndromes patients, particularly in those with high pre-therapy EVI1 expression|url=https://www.ncbi.nlm.nih.gov/pubmed/15203277|journal=Leukemia Research|volume=28|issue=8|pages=791–803|doi=10.1016/j.leukres.2003.11.018|issn=0145-2126|pmid=15203277}}</ref>. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Individual Region Genomic Gain / Loss / LOH== | ==Individual Region Genomic Gain / Loss / LOH== | ||
| Line 198: | Line 204: | ||
|} | |} | ||
<blockquote class='blockedit'>{{Box-round|title=v4:Genomic Gain/Loss/LOH|The content below was from the old template. Please incorporate above.}} | <blockquote class='blockedit'>{{Box-round|title=v4:Genomic Gain/Loss/LOH|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
Put your text here. | Put your text here. | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Characteristic Chromosomal Patterns== | ==Characteristic Chromosomal Patterns== | ||
| Line 225: | Line 234: | ||
|} | |} | ||
<blockquote class='blockedit'>{{Box-round|title=v4:Characteristic Chromosomal Aberrations / Patterns|The content below was from the old template. Please incorporate above.}} | <blockquote class='blockedit'>{{Box-round|title=v4:Characteristic Chromosomal Aberrations / Patterns|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
Monosomy 7 is the most common associated (secondary) cytogenetic abnormality (in ~66% of cases), and appears to further contribute to the adverse prognosis of the inv(3q)/t(3;3) abnormality<ref name=":0" /><ref name=":1" />. | Monosomy 7 is the most common associated (secondary) cytogenetic abnormality (in ~66% of cases), and appears to further contribute to the adverse prognosis of the inv(3q)/t(3;3) abnormality<ref name=":0" /><ref name=":1" />. | ||
| Line 231: | Line 240: | ||
Deletion 5q | Deletion 5q | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Gene Mutations (SNV / INDEL)== | ==Gene Mutations (SNV / INDEL)== | ||
| Line 266: | Line 278: | ||
<blockquote class='blockedit'>{{Box-round|title=v4:Gene Mutations (SNV/INDEL)|The content below was from the old template. Please incorporate above.}} | <blockquote class='blockedit'>{{Box-round|title=v4:Gene Mutations (SNV/INDEL)|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
Secondary mutations are found in all AML cases with inv(3) ot t(3;3). Mutations in genes activating RAS/TK signaling pathway are the most common mutation. | Secondary mutations are found in all AML cases with inv(3) ot t(3;3). Mutations in genes activating RAS/TK signaling pathway are the most common mutation. | ||
| Line 283: | Line 295: | ||
|} | |} | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Epigenomic Alterations== | ==Epigenomic Alterations== | ||
| Line 308: | Line 323: | ||
|} | |} | ||
<blockquote class='blockedit'>{{Box-round|title=v4:Genes and Main Pathways Involved|The content below was from the old template. Please incorporate above.}} | <blockquote class='blockedit'>{{Box-round|title=v4:Genes and Main Pathways Involved|The content below was from the old template. Please incorporate above.}}</blockquote> | ||
The ''[[MECOM]]'' (MDS1 and EVI1 complex locus) gene in 3q26.3 is the key gene implicated in pathogenesis of AML with inv(3)/t(3;3). ''MECOM'' codes for several differentially spliced transcripts: MDS1-EVI1, MDS1 and EVI1. It consequently yields the MDS1-EVI1 [1239 amino acids (AA)], MDS1 (188AA) and EVI1 (1051AA) protein isoforms. While ''EVI1'' deregulated expression has been reported to be critical in stem cell self-renewal and leukaemogenesis, the roles of ''MDS1'' and MDS1-EVI1 in haematological malignancies remain unclear<ref name=":2">{{Cite journal|last=Wieser|first=Rotraud|date=2007|title=The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions|url=https://www.ncbi.nlm.nih.gov/pubmed/17507183|journal=Gene|volume=396|issue=2|pages=346–357|doi=10.1016/j.gene.2007.04.012|issn=0378-1119|pmid=17507183}}</ref>. | The ''[[MECOM]]'' (MDS1 and EVI1 complex locus) gene in 3q26.3 is the key gene implicated in pathogenesis of AML with inv(3)/t(3;3). ''MECOM'' codes for several differentially spliced transcripts: MDS1-EVI1, MDS1 and EVI1. It consequently yields the MDS1-EVI1 [1239 amino acids (AA)], MDS1 (188AA) and EVI1 (1051AA) protein isoforms. While ''EVI1'' deregulated expression has been reported to be critical in stem cell self-renewal and leukaemogenesis, the roles of ''MDS1'' and MDS1-EVI1 in haematological malignancies remain unclear<ref name=":2">{{Cite journal|last=Wieser|first=Rotraud|date=2007|title=The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions|url=https://www.ncbi.nlm.nih.gov/pubmed/17507183|journal=Gene|volume=396|issue=2|pages=346–357|doi=10.1016/j.gene.2007.04.012|issn=0378-1119|pmid=17507183}}</ref>. | ||
| Line 322: | Line 337: | ||
Key cellular pathways: ''EVI1'' has been shown to be involved in multiple downstream signaling pathways, including the transforming growth factor beta (TGF-β) signaling (where ''EVI1'' prevents transcription of the TGF-β induced anti-growth genes ) and ''[[JUN|c-Jun N-terminal kinase]]'' (JNK) signaling (where ''EVI1'' prevents phosphorylation and activation of key transcription factors for the apoptotic response). | Key cellular pathways: ''EVI1'' has been shown to be involved in multiple downstream signaling pathways, including the transforming growth factor beta (TGF-β) signaling (where ''EVI1'' prevents transcription of the TGF-β induced anti-growth genes ) and ''[[JUN|c-Jun N-terminal kinase]]'' (JNK) signaling (where ''EVI1'' prevents phosphorylation and activation of key transcription factors for the apoptotic response). | ||
<blockquote class="blockedit"> | |||
<center><span style="color:Maroon">'''End of V4 Section'''</span> | |||
---- | |||
</blockquote> | </blockquote> | ||
==Genetic Diagnostic Testing Methods== | ==Genetic Diagnostic Testing Methods== | ||
Revision as of 13:10, 10 February 2025
Haematolymphoid Tumours (WHO Classification, 5th ed.)
| This page is under construction |
editContent Update To WHO 5th Edition Classification Is In Process; Content Below is Based on WHO 4th Edition ClassificationThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Acute Myeloid Leukemia (AML) with inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2);GATA2, MECOM.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Gordana Raca MD PhD, University of Southern California, Los Angeles
WHO Classification of Disease
| Structure | Disease |
|---|---|
| Book | Haematolymphoid Tumours (5th ed.) |
| Category | Myeloid proliferations and neoplasms |
| Family | Acute myeloid leukaemia |
| Type | Acute myeloid leukaemia with defining genetic abnormalities |
| Subtype(s) | Acute myeloid leukaemia with MECOM rearrangement |
Definition / Description of Disease
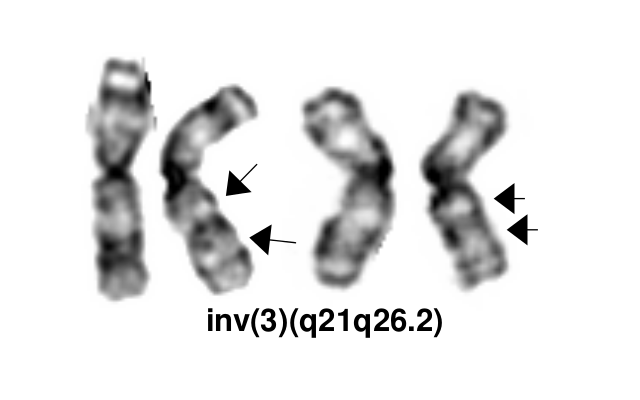
The inv(3)(q21q26.2)/t(3;3)(q21;q26.2) defines a distinctive cytogenetic subtype of acute myeloid leukemia (AML) in 2008 WHO classification (AML with inv(3)(q21q26.2)/t(3;3)(q21;q26.2), although it is currently not recognized as an abnormality pathognomonic for diagnosis of AML irrespective of blast percentage. This abnormality also occurs in myelodysplastic syndrome (MDS), where it is associated with aggressive course and a high risk of progression to AML. It can also rarely be found in other myeloid neoplasms, including chronic myelogenous leukemia (mostly during accelerated phase or in blast crisis), other myeloproliferative neoplasms (MPN), and in overlap MDS/MPNs including chronic myelomonocytic leukemia[1].
inv(3)(q21q26.2) and t(3;3)(q21;q26.2) are variant rearrangements with similar consequences at the molecular level and similar disease associations and clinical implications.
In addition to the balanced inversion and translocation involving the bands 3q21 and 3q26.2, other 3q abnormalities are also common in AML. A study of 6515 adults with newly diagnosed AML detected 3q abnormalities in 4.4% of the patients, and classified them in four categories[2]:
- t(3;3)/inv(3)
- Translocations involving only the band 3q26.2
- Translocations involving only the band 3q21
- Other 3q abnormalities.
Further studies are needed to determine how the molecular pathogenesis and clinical features of AML with other 3q rearrangements compare with the AML with inv(3)/t(3;3).
Synonyms / Terminology
AML with RPN1-MECOM
Epidemiology / Prevalence
AML with inv(3)(q21q26.2) and t(3;3)(q21;q26.2) comprises 1-2.5% of all AML cases and less than 1% of all MDS cases[1][2].
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Fatigue EXAMPLE: Lymphadenopathy (uncommon) |
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE: Lymphocytosis (low level) |
editv4:Clinical FeaturesThe content below was from the old template. Please incorporate above.
The patients often present with anemia and normal or increased platelet counts. However, decreased platelet counts and hepatosplenomegaly can also be observed[1][2].
End of V4 Section
Sites of Involvement
Bone Marrow.
Morphologic Features
Common morphologic features include multilineage dysplasia, with atypical megakaryocytes (ie, small mono- or bilobed forms) and variable fibrosis. The most consistent bone marrow finding in this disorder is an increase in the number of megakaryocytes, many of which are morphologically abnormal. In some patients, almost all of the identifiable megakaryocytes are micromegakaryocytes.
Immunophenotype
Immunophenotypic studies typically show expression of CD13, CD33, HLA-DR, CD17(KIT) CD34 and CD38, occasionally with aberrant expression of CD7 and megakaryocytic markers like CD41 and CD61.
| Finding | Marker |
|---|---|
| Positive (universal) | EXAMPLE: CD1 |
| Positive (subset) | EXAMPLE: CD2 |
| Negative (universal) | EXAMPLE: CD3 |
| Negative (subset) | EXAMPLE: CD4 |
Chromosomal Rearrangements (Gene Fusions)
Put your text here and fill in the table
| Chromosomal Rearrangement | Genes in Fusion (5’ or 3’ Segments) | Pathogenic Derivative | Prevalence | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE: t(9;22)(q34;q11.2) | EXAMPLE: 3'ABL1 / 5'BCR | EXAMPLE: der(22) | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add reference) |
Yes | No | Yes | EXAMPLE:
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). |
editv4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).Please incorporate this section into the relevant tables found in:
- Chromosomal Rearrangements (Gene Fusions)
- Individual Region Genomic Gain/Loss/LOH
- Characteristic Chromosomal Patterns
- Gene Mutations (SNV/INDEL)
Diagnostic: The presence of inv(3)/t(3;3) defines a cytogenetic subtype of AML, however in the current WHO 2017 classification does not allow to make a diagnosis of AML if blast percentage <20%. Very poor prognosis and rapid progression of MDS with inv(3)(q21q26.2)/t(3;3)(q21;q26.2) resulted in a proposal to consider neoplasms with these rearrangements as an AML with recurrent genetic abnormalities, irrespective of blast percentage.
BCR-ABL1 positive patients with CML and concomitant MECOM rearrangement are considered in an aggressive phase of CML (Accelerated Phase) rather than de novo AML with inv(3)/t(3;3)
Prognostic: The inv(3)(q21q26.2)/t(3;3)(q21;q26.2) is associated with a dismal prognosis in both AML and MDS. Review of 6515 adult patients with newly diagnosed AML enrolled in prospective European trials showed 5-year OS of only 5.7% +/- 3 for AML with inv(3)/t(3;3), with the median survival os 10.3 months. This adverse prognostic impact of inv(3)/t(3;3) appears to be further enhanced by additional monosomy 7 and/or complex karyotype. Similarly, the prognosis of MDS with inv(3)/t(3;3) is also poor, and the revised International Prognostic Scoring System (IPSS-R) for MDS includes inv(3)/t(3;3) among its poor risk cytogenetic abnormalities[1][2].
Therapeutic: AML with inv(3)(q21q26.2) or t(3;3) (q21;q26.2) is an aggressive disease with short survival, in need for novel and more efficient treatments. Some studies have shown that arsenic trioxide induces targeted specific degradation of the AML1/MDS1/EVI1 oncoprotein, and an isolated case report described good response to Arsenic trioxide and thalidomide combination in a patient with MDS with inv(3)[3][4].
End of V4 Section
Individual Region Genomic Gain / Loss / LOH
Put your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
| Chr # | Gain / Loss / Amp / LOH | Minimal Region Genomic Coordinates [Genome Build] | Minimal Region Cytoband | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|
| EXAMPLE:
7 |
EXAMPLE: Loss | EXAMPLE:
chr7:1- 159,335,973 [hg38] |
EXAMPLE:
chr7 |
Yes | Yes | No | EXAMPLE:
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). |
| EXAMPLE:
8 |
EXAMPLE: Gain | EXAMPLE:
chr8:1-145,138,636 [hg38] |
EXAMPLE:
chr8 |
No | No | No | EXAMPLE:
Common recurrent secondary finding for t(8;21) (add reference). |
editv4:Genomic Gain/Loss/LOHThe content below was from the old template. Please incorporate above.
Put your text here.
End of V4 Section
Characteristic Chromosomal Patterns
Put your text here (EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.)
| Chromosomal Pattern | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|
| EXAMPLE:
Co-deletion of 1p and 18q |
Yes | No | No | EXAMPLE:
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). |
editv4:Characteristic Chromosomal Aberrations / PatternsThe content below was from the old template. Please incorporate above.
Monosomy 7 is the most common associated (secondary) cytogenetic abnormality (in ~66% of cases), and appears to further contribute to the adverse prognosis of the inv(3q)/t(3;3) abnormality[1][2]. Complex karyotypes Deletion 5q
End of V4 Section
Gene Mutations (SNV / INDEL)
Put your text here and fill in the table (Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well as either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable. Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.)
| Gene; Genetic Alteration | Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other) | Prevalence (COSMIC / TCGA / Other) | Concomitant Mutations | Mutually Exclusive Mutations | Diagnostic Significance (Yes, No or Unknown) | Prognostic Significance (Yes, No or Unknown) | Therapeutic Significance (Yes, No or Unknown) | Notes |
|---|---|---|---|---|---|---|---|---|
| EXAMPLE: TP53; Variable LOF mutations
EXAMPLE: EGFR; Exon 20 mutations EXAMPLE: BRAF; Activating mutations |
EXAMPLE: TSG | EXAMPLE: 20% (COSMIC)
EXAMPLE: 30% (add Reference) |
EXAMPLE: IDH1 R123H | EXAMPLE: EGFR amplification | EXAMPLE: Excludes hairy cell leukemia (HCL) (add reference).
|
Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content.
editv4:Gene Mutations (SNV/INDEL)The content below was from the old template. Please incorporate above.
Secondary mutations are found in all AML cases with inv(3) ot t(3;3). Mutations in genes activating RAS/TK signaling pathway are the most common mutation.
NRAS (~30%), PTPN11(11%), FLT3 (13%) and KRAS (11%), as well as GATA2 (15%) , RUNX1 (12%) and NF1 (9%) , CBL (7%) , KIT (2%)[5]. CBL mutation is found often with concomitant GATA2 mutation.
Other Mutations
| Type | Gene/Region/Other |
|---|---|
| Concomitant Mutations | GATA2, CBL |
| Secondary Mutations | |
| Mutually Exclusive |
End of V4 Section
Epigenomic Alterations
Put your text here
Genes and Main Pathways Involved
Put your text here and fill in the table (Instructions: Can include references in the table. Do not delete table.)
| Gene; Genetic Alteration | Pathway | Pathophysiologic Outcome |
|---|---|---|
| EXAMPLE: BRAF and MAP2K1; Activating mutations | EXAMPLE: MAPK signaling | EXAMPLE: Increased cell growth and proliferation |
| EXAMPLE: CDKN2A; Inactivating mutations | EXAMPLE: Cell cycle regulation | EXAMPLE: Unregulated cell division |
| EXAMPLE: KMT2C and ARID1A; Inactivating mutations | EXAMPLE: Histone modification, chromatin remodeling | EXAMPLE: Abnormal gene expression program |
editv4:Genes and Main Pathways InvolvedThe content below was from the old template. Please incorporate above.
The MECOM (MDS1 and EVI1 complex locus) gene in 3q26.3 is the key gene implicated in pathogenesis of AML with inv(3)/t(3;3). MECOM codes for several differentially spliced transcripts: MDS1-EVI1, MDS1 and EVI1. It consequently yields the MDS1-EVI1 [1239 amino acids (AA)], MDS1 (188AA) and EVI1 (1051AA) protein isoforms. While EVI1 deregulated expression has been reported to be critical in stem cell self-renewal and leukaemogenesis, the roles of MDS1 and MDS1-EVI1 in haematological malignancies remain unclear[6].
EVI1 is a zinc-finger nuclear transcription factor involved in many signaling pathways. It binds DNA through specific conserved sequences, interacts with a number of transcriptional and epigenetic regulators (CREBBP, CTBP, HDAC, KAT2B (P/CAF), SMAD3, GATA1, GATA2, DNMT3A, and DNMT3B), and mediates chromatin modifications and DNA hypermethylation. EVI1 plays a role in embryogenesis and its deficiency in mice is an embryonic lethal mutation. Among the many other observed defects, EVI1-/- mouse embryos have been shown to have defects in both the development and proliferation of hematopoietic stem cells (HSCs). It appears that depending on its binding partners, EVI1 can act as a transcriptional activator to promote the proliferation of hematopoietic stem cells (eg, when bound to GATA2) or as a transcriptional repressor inhibiting erythroid differentiation (eg, when bound to GATA1)[6].
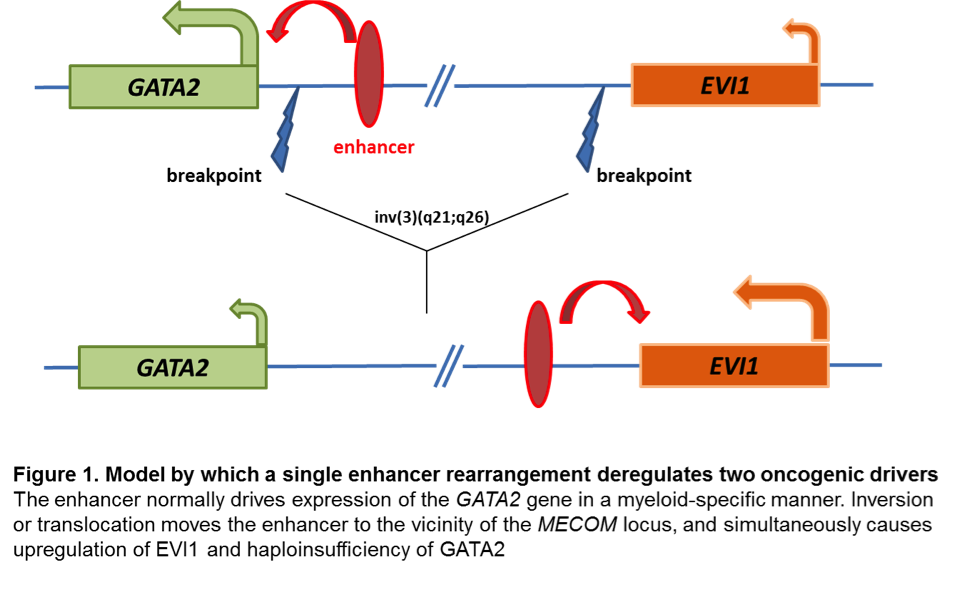
Changes at mRNA level: EVI1 proto-oncogene has early on been identified as aberrantly upregulated in almost all AML cases with inv(3) and t(3;3). Aberrant EVI1 expression is also found in a majority of AML patients with other 3q26 abnormalities and even in patients with myeloid leukemia and a normal karyotype, suggesting that inappropriate activation of this gene occurs through various mechanisms[7][8][9], but invariably confers an adverse prognosis. Interestingly, the breakpoints in chromosomal rearrangements that lead to EVI1 activation in AML with inv(3)/t(3;3) were shown to occur either 5' of the gene in the t(3;3) or 3' of the gene in the inv(3)[7]. It has long been thought that activation of EVI1 in malignant hematopoietic cells occurs by juxtaposition of the gene to enhancer elements of the housekeeping ribophorin gene located at 3q21. However, several groups have recently described a novel molecular mechanism by which the inv(3)/t(3;3) re-positions a distal GATA2 enhancer to activate MECOM expression, and simultaneously confer GATA2 functional haploinsufficiency[10][11]. This mechanism presents a new paradigm for the pathogenesis of MDS/AML.
Changes at protein level: The inv(3)(q21q26.2)/t(3;3)(q21;q26.2) results in upregulation of the MECOM gene and increased levels of the EVI1 protein, but does not produce abnormal gene and protein fusions.
Key cellular pathways: EVI1 has been shown to be involved in multiple downstream signaling pathways, including the transforming growth factor beta (TGF-β) signaling (where EVI1 prevents transcription of the TGF-β induced anti-growth genes ) and c-Jun N-terminal kinase (JNK) signaling (where EVI1 prevents phosphorylation and activation of key transcription factors for the apoptotic response).
End of V4 Section
Genetic Diagnostic Testing Methods
- Karyotype: Inv(3) and t(3;3) can typically be detected by routine karyotype analysis, although the inversion may be challenging to identify in the cases with poor morphology.
- FISH: EVI1 break-apart FISH probe is commercially available and FISH testing is offered clinically by most clinical cytogenetic laboratories.
Familial Forms
Put your text here (Instructions: Include associated hereditary conditions/syndromes that cause this entity or are caused by this entity.)
Additional Information
Put your text here
Links
Up To Date Cytogenetics in AML
References
(use the "Cite" icon at the top of the page) (Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted. If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference.)
- ↑ 1.0 1.1 1.2 1.3 1.4 Rogers, Heesun J.; et al. (2014). "Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv(3)(q21q26.2)/t(3;3)(q21;q26.2): a Bone Marrow Pathology Group study". Haematologica. 99 (5): 821–829. doi:10.3324/haematol.2013.096420. ISSN 1592-8721. PMC 4008101. PMID 24463215.
- ↑ 2.0 2.1 2.2 2.3 2.4 Lugthart, Sanne; et al. (2010). "Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia". Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 28 (24): 3890–3898. doi:10.1200/JCO.2010.29.2771. ISSN 1527-7755. PMID 20660833.
- ↑ Shackelford, David; et al. (2006). "Targeted degradation of the AML1/MDS1/EVI1 oncoprotein by arsenic trioxide". Cancer Research. 66 (23): 11360–11369. doi:10.1158/0008-5472.CAN-06-1774. ISSN 0008-5472. PMID 17145882.
- ↑ Raza, Azra; et al. (2004). "Arsenic trioxide and thalidomide combination produces multi-lineage hematological responses in myelodysplastic syndromes patients, particularly in those with high pre-therapy EVI1 expression". Leukemia Research. 28 (8): 791–803. doi:10.1016/j.leukres.2003.11.018. ISSN 0145-2126. PMID 15203277.
- ↑ Haferlach, C.; et al. (2011). "The inv(3)(q21q26)/t(3;3)(q21;q26) is frequently accompanied by alterations of the RUNX1, KRAS and NRAS and NF1 genes and mediates adverse prognosis both in MDS and in AML: a study in 39 cases of MDS or AML". Leukemia. 25 (5): 874–877. doi:10.1038/leu.2011.5. ISSN 1476-5551. PMID 21283084.
- ↑ 6.0 6.1 Wieser, Rotraud (2007). "The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions". Gene. 396 (2): 346–357. doi:10.1016/j.gene.2007.04.012. ISSN 0378-1119. PMID 17507183.
- ↑ 7.0 7.1 Morishita, K.; et al. (1992). "Activation of EVI1 gene expression in human acute myelogenous leukemias by translocations spanning 300-400 kilobases on chromosome band 3q26". Proceedings of the National Academy of Sciences of the United States of America. 89 (9): 3937–3941. doi:10.1073/pnas.89.9.3937. ISSN 0027-8424. PMC 525606. PMID 1570317.CS1 maint: PMC format (link)
- ↑ Russell, M.; et al. (1994). "Expression of EVI1 in myelodysplastic syndromes and other hematologic malignancies without 3q26 translocations". Blood. 84 (4): 1243–1248. ISSN 0006-4971. PMID 8049440.
- ↑ Langabeer, S. E.; et al. (2001). "EVI1 expression in acute myeloid leukaemia". British Journal of Haematology. 112 (1): 208–211. doi:10.1046/j.1365-2141.2001.02569.x. ISSN 0007-1048. PMID 11167805.
- ↑ Gröschel, Stefan; et al. (2014). "A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia". Cell. 157 (2): 369–381. doi:10.1016/j.cell.2014.02.019. ISSN 1097-4172. PMID 24703711.
- ↑ Yamazaki, Hiromi; et al. (2014). "A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression". Cancer Cell. 25 (4): 415–427. doi:10.1016/j.ccr.2014.02.008. ISSN 1878-3686. PMC 4012341. PMID 24703906.
Notes
*Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome.
Edited by: Fabiola Quintero-Rivera 8/3/2018
*Citation of this Page: “Acute myeloid leukaemia with MECOM rearrangement”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated 02/10/2025, https://ccga.io/index.php/HAEM5:Acute_myeloid_leukaemia_with_MECOM_rearrangement.